NF1) Inactivation Characterizes NF1-Associated Pilocytic Astrocytoma
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Leptomeningeal Dissemination of Pilocytic Astrocytoma Via Hematoma in a Child
Neurosurg Focus 13 (1):Clinical Pearl 2, 2002, Click here to return to Table of Contents Leptomeningeal dissemination of pilocytic astrocytoma via hematoma in a child Case report MASARU KANDA, M.D., HIDENOBU TANAKA, M.D., PH.D., SOJI SHINODA, M.D., PH.D., AND TOSHIO MASUZAWA, M.D., PH.D. Department of Surgical Neurology, Jichi Medical School, Tochigi, Japan A case of recurrent pilocytic astrocytoma with leptomeningeal dissemination (LMD) is described. A cerebellar tumor was diagnosed in a 3-year-old boy, in whom resection was performed. When the boy was 6 years of age, recur- rence was treated with surgery and local radiotherapy. At age 13 years, scoliosis was present, but the patient was asymptomatic. Twelve years after initial surgery LMD was demonstrated in the lumbar spinal region without recur- rence of the original tumor. This tumor also was subtotally removed. During the procedure, a hematoma was observed adjacent to the tumor, but the border was clear. Histological examination of the spinal cord tumor showed features sim- ilar to those of the original tumor. There were no tumor cells in the hematoma. The MIB-1 labeling index indicated no malignant change compared with the previous samples. Radiotherapy was performed after the surgery. The importance of early diagnosis and management of scoliosis is emphasized, and the peculiar pattern of dissemination of the pilo- cytic astrocytoma and its treatment are reviewed. KEY WORDS • pilocytic astrocytoma • leptomeningeal dissemination • MIB-1 labeling index • radiation therapy • scoliosis -

Malignant Peripheral Nerve Sheath Tumor (MPNST): an Overview with Emphasis on Pathology, Imaging and Management Strategies
Thomas Jefferson University Jefferson Digital Commons Department of Neurosurgery Faculty Papers Department of Neurosurgery 6-2012 Malignant Peripheral Nerve Sheath Tumor (MPNST): An overview with emphasis on pathology, imaging and management strategies Timothy C. Beer 3rd year medical student, Jefferson Medical College Follow this and additional works at: https://jdc.jefferson.edu/neurosurgeryfp Part of the Medical Neurobiology Commons Let us know how access to this document benefits ouy Recommended Citation Beer, Timothy C., "Malignant Peripheral Nerve Sheath Tumor (MPNST): An overview with emphasis on pathology, imaging and management strategies" (2012). Department of Neurosurgery Faculty Papers. Paper 18. https://jdc.jefferson.edu/neurosurgeryfp/18 This Article is brought to you for free and open access by the Jefferson Digital Commons. The Jefferson Digital Commons is a service of Thomas Jefferson University's Center for Teaching and Learning (CTL). The Commons is a showcase for Jefferson books and journals, peer-reviewed scholarly publications, unique historical collections from the University archives, and teaching tools. The Jefferson Digital Commons allows researchers and interested readers anywhere in the world to learn about and keep up to date with Jefferson scholarship. This article has been accepted for inclusion in Department of Neurosurgery Faculty Papers by an authorized administrator of the Jefferson Digital Commons. For more information, please contact: [email protected]. An overview with emphasis -

Central Nervous System
PRINCESS MARGARET CANCER CENTRE CLINICAL PRACTICE GUIDELINES CENTRAL NERVOUS SYSTEM LOW GRADE GLIOMAS CNS Site Group – Low Grade Gliomas Author: Dr. Norm Laperriere 1. INTRODUCTION 3 2. PREVENTION 3 3. SCREENING AND EARLY DETECTION 3 4. DIAGNOSIS AND PATHOLOGY 3 5. MANAGEMENT 4 5.1 MANAGEMENT ALGORITHMS 4 5.2 SURGERY 4 5.3 CHEMOTHERAPY 5 5.4 RADIATION THERAPY 5 6. ONCOLOGY NURSING PRACTICE 6 7. SUPPORTIVE CARE 6 7.1 PATIENT EDUCATION 6 7.2 PSYCHOSOCIAL CARE 6 7.3 SYMPTOM MANAGEMENT 6 7.4 CLINICAL NUTRITION 7 7.5 PALLIATIVE CARE 7 7.6 REHABILITATION 7 8. FOLLOW-UP CARE 7 Last Revision Date – April 2019 2 Low Grade Gliomas 1. Introduction • Grade I gliomas: pilocytic astrocytoma (PA), dysembryoplastic neuroepithelial tumor (DNET), pleomorphic xanthoastrocytoma, (PXA), ganglioglioma • Grade II gliomas: infiltrating astrocytoma, oligodendroglioma, mixed gliomas • annual incidence is approx. 1/100,000 This document is intended for use by members of the Central Nervous System site group of the Princess Margaret Hospital/University Health Network. The guidelines in this document are meant as a guide only, and are not meant to be prescriptive. There exists a multitude of individual factors, prognostic factors and peculiarities in any individual case, and for that reason the ultimate decision as to the management of any individual patient is at the discretion of the staff physician in charge of that particular patient’s care. 2. Prevention • genetic counseling for all NF1 carriers 3. Screening and Early Detection • baseline MRI brain for all newly -

Central Nervous System Tumors General ~1% of Tumors in Adults, but ~25% of Malignancies in Children (Only 2Nd to Leukemia)
Last updated: 3/4/2021 Prepared by Kurt Schaberg Central Nervous System Tumors General ~1% of tumors in adults, but ~25% of malignancies in children (only 2nd to leukemia). Significant increase in incidence in primary brain tumors in elderly. Metastases to the brain far outnumber primary CNS tumors→ multiple cerebral tumors. One can develop a very good DDX by just location, age, and imaging. Differential Diagnosis by clinical information: Location Pediatric/Young Adult Older Adult Cerebral/ Ganglioglioma, DNET, PXA, Glioblastoma Multiforme (GBM) Supratentorial Ependymoma, AT/RT Infiltrating Astrocytoma (grades II-III), CNS Embryonal Neoplasms Oligodendroglioma, Metastases, Lymphoma, Infection Cerebellar/ PA, Medulloblastoma, Ependymoma, Metastases, Hemangioblastoma, Infratentorial/ Choroid plexus papilloma, AT/RT Choroid plexus papilloma, Subependymoma Fourth ventricle Brainstem PA, DMG Astrocytoma, Glioblastoma, DMG, Metastases Spinal cord Ependymoma, PA, DMG, MPE, Drop Ependymoma, Astrocytoma, DMG, MPE (filum), (intramedullary) metastases Paraganglioma (filum), Spinal cord Meningioma, Schwannoma, Schwannoma, Meningioma, (extramedullary) Metastases, Melanocytoma/melanoma Melanocytoma/melanoma, MPNST Spinal cord Bone tumor, Meningioma, Abscess, Herniated disk, Lymphoma, Abscess, (extradural) Vascular malformation, Metastases, Extra-axial/Dural/ Leukemia/lymphoma, Ewing Sarcoma, Meningioma, SFT, Metastases, Lymphoma, Leptomeningeal Rhabdomyosarcoma, Disseminated medulloblastoma, DLGNT, Sellar/infundibular Pituitary adenoma, Pituitary adenoma, -

Differentiation Between Pilocytic Astrocytoma and Glioblastoma: a Decision Tree Model Using Contrast-Enhanced Magnetic Resonance
European Radiology (2019) 29:3968–3975 https://doi.org/10.1007/s00330-018-5706-6 ONCOLOGY Differentiation between pilocytic astrocytoma and glioblastoma: a decision tree model using contrast-enhanced magnetic resonance imaging-derived quantitative radiomic features Fei Dong1 & Qian Li1 & Duo Xu1 & Wenji Xiu2 & Qiang Zeng3 & Xiuliang Zhu1 & Fangfang Xu1 & Biao Jiang1 & Minming Zhang1 Received: 13 March 2018 /Revised: 8 July 2018 /Accepted: 6 August 2018 /Published online: 12 November 2018 # European Society of Radiology 2018 Abstract Objective To differentiate brain pilocytic astrocytoma (PA) from glioblastoma (GBM) using contrast-enhanced mag- netic resonance imaging (MRI) quantitative radiomic features by a decision tree model. Methods Sixty-six patients from two centres (PA, n = 31; GBM, n = 35) were randomly divided into training and validation data sets (about 2:1). Quantitative radiomic features of the tumours were extracted from contrast-enhanced MR images. A subset of features was selected by feature stability and Boruta algorithm. The selected features were used to build a decision tree model. Predictive accuracy, sensitivity and specificity were used to assess model performance. The classification outcome of the model was combined with tumour location, age and gender features, and multivariable logistic regression analysis and permutation test using the entire data set were performed to further evaluate the decision tree model. Results A total of 271 radiomic features were successfully extracted for each tumour. Twelve features were selected as input variables to build the decision tree model. Two features S(1, -1) Entropy and S(2, -2) SumAverg were finally included in the model. The model showed an accuracy, sensitivity and specificity of 0.87, 0.90 and 0.83 for the training data set and 0.86, 0.80 and 0.91 for the validation data set. -
A Case of Intramedullary Spinal Cord Astrocytoma Associated with Neurofibromatosis Type 1
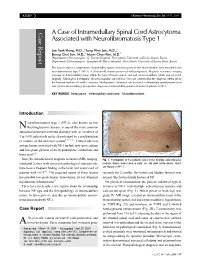
KISEP J Korean Neurosurg Soc 36 : 69-71, 2004 Case Report A Case of Intramedullary Spinal Cord Astrocytoma Associated with Neurofibromatosis Type 1 Jae Taek Hong, M.D.,1 Sang Won Lee, M.D.,1 Byung Chul Son, M.D.,1 Moon Chan Kim, M.D.2 Department of Neurosurgery,1 St. Vincent Hospital, The Catholic University of Korea, Suwon, Korea Department of Neurosurgery,2 Kangnam St. Mary's Hospital, The Catholic University of Korea, Seoul, Korea The authors report a symptomatic intramedullary spinal cord astrocytoma in the thoracolumbar area associated with neurofibromatosis type 1 (NF-1). A 38-year-old woman presented with paraparesis. Magnetic resonance imaging revealed an intramedullary lesion within the lower thoracic spinal cord and conus medullaris, which was removed surgically. Pathological investigation showed anaplastic astrocytoma. This case confirms that the diagnosis criteria set by the National Institute of Health Consensus Development Conference can be useful to differentiate ependymoma from astrocytoma when making a preoperative diagnosis of intramedullary spinal cord tumor in patients of NF-1. KEY WORDS : Astrocytoma·Intramedullary cord tumor·Neurofibromatosis. Introduction eurofibromatosis type 1 (NF-1), also known as von N Recklinghausen's disease, is one of the most common autosomal dominant inherited disorders with an incidence of 1 in 3,000 individuals and is characterized by a predisposition to tumors of the nervous system5,6,12,16). Central nervous system lesions associated with NF-1 include optic nerve glioma and low-grade gliomas of the hypothalamus, cerebellum and brain stem6,10). Since the introduction of magnetic resonance(MR) imaging, Fig. 1. Photograph of the patient's back shows multiple subcutaneous incidental lesions with uncertain pathological characteristic nodules (black arrow) and a cafe-au-lait spot (white arrow), which have been a frequent finding in the brain and spinal cord of are typical of NF-1. -

Pearls and Forget-Me-Nots in the Management of Retinoblastoma
POSTERIOR SEGMENT ONCOLOGY FEATURE STORY Pearls and Forget-Me-Nots in the Management of Retinoblastoma Retinoblastoma represents approximately 4% of all pediatric malignancies and is the most common intraocular malignancy in children. BY CAROL L. SHIELDS, MD he management of retinoblastoma has gradu- ular malignancy in children.1-3 It is estimated that 250 to ally evolved over the years from enucleation to 300 new cases of retinoblastoma are diagnosed in the radiotherapy to current techniques of United States each year, and 5,000 cases are found world- chemotherapy. Eyes with massive retinoblas- Ttoma filling the globe are still managed with enucleation, TABLE 1. INTERNATIONAL CLASSIFICATION OF whereas those with small, medium, or even large tumors RETINOBLASTOMA (ICRB) can be managed with chemoreduction followed by Group Quick Reference Specific Features tumor consolidation with thermotherapy or cryotherapy. A Small tumor Rb <3 mm* Despite multiple or large tumors, visual acuity can reach B Larger tumor Rb >3 mm* or ≥20/40 in many cases, particularly in eyes with extrafoveal retinopathy, and facial deformities that have Macula Macular Rb location been found following external beam radiotherapy are not (<3 mm to foveola) anticipated following chemoreduction. Recurrence from Juxtapapillary Juxtapapillary Rb location subretinal and vitreous seeds can be problematic. Long- (<1.5 mm to disc) term follow-up for second cancers is advised. Subretinal fluid Rb with subretinal fluid Most of us can only remember a few interesting points C Focal seeds Rb with: from a lecture, even if was delivered by an outstanding, Subretinal seeds <3 mm from Rb colorful speaker. Likewise, we generally retain only a small and/or percentage of the information that we read, even if writ- Vitreous seeds <3 mm ten by the most descriptive or lucent author. -

UCSD Moores Cancer Center Neuro-Oncology Program
UCSD Moores Cancer Center Neuro-Oncology Program Recent Progress in Brain Tumors 6DQWRVK.HVDUL0'3K' 'LUHFWRU1HXUR2QFRORJ\ 3URIHVVRURI1HXURVFLHQFHV 0RRUHV&DQFHU&HQWHU 8QLYHUVLW\RI&DOLIRUQLD6DQ'LHJR “Brain Cancer for Life” Juvenile Pilocytic Astrocytoma Metastatic Brain Cancer Glioblastoma Multiforme Glioblastoma Multiforme Desmoplastic Infantile Ganglioglioma Desmoplastic Variant Astrocytoma Medulloblastoma Atypical Teratoid Rhabdoid Tumor Diffuse Intrinsic Pontine Glioma -Mutational analysis, microarray expression, epigenetic phenomenology -Age-specific biology of brain cancer -Is there an overlap? ? Neuroimmunology ? Stem cell hypothesis Courtesy of Dr. John Crawford Late Effects Long term effect of chemotherapy and radiation on neurocognition Risks of secondary malignancy secondary to chemotherapy and/or radiation Neurovascular long term effects: stroke, moya moya Courtesy of Dr. John Crawford Importance Increase in aging population with increased incidence of cancer Patients with cancer living longer and developing neurologic disorders due to nervous system relapse or toxicity from treatments Overview Introduction Clinical Presentation Primary Brain Tumors Metastatic Brain Tumors Leptomeningeal Metastases Primary CNS Lymphoma Paraneoplastic Syndromes Classification of Brain Tumors Tumors of Neuroepithelial Tissue Glial tumors (astrocytic, oligodendroglial, mixed) Neuronal and mixed neuronal-glial tumors Neuroblastic tumors Pineal parenchymal tumors Embryonal tumors Tumors of Peripheral Nerves Shwannoma Neurofibroma -

Metabolic Profiling of the Three Neural Derived Embryonal Pediatric Tumors
Metabolic profiling of the three neural derived embryonal pediatric tumors retinoblastoma, neuroblastoma and medulloblastoma, identifies distinct metabolic profiles Kohe, Sarah; Bennett, Christopher; Gill, Simrandip; Wilson, Martin; McConville, Carmel; Peet, Andrew DOI: 10.18632/oncotarget.24168 License: Creative Commons: Attribution (CC BY) Document Version Publisher's PDF, also known as Version of record Citation for published version (Harvard): Kohe, SE, Bennett, CD, Gill, SK, Wilson, M, McConville, C & Peet, AC 2018, 'Metabolic profiling of the three neural derived embryonal pediatric tumors retinoblastoma, neuroblastoma and medulloblastoma, identifies distinct metabolic profiles', OncoTarget, vol. 9, no. 13, pp. 11336-11351. https://doi.org/10.18632/oncotarget.24168 Link to publication on Research at Birmingham portal General rights Unless a licence is specified above, all rights (including copyright and moral rights) in this document are retained by the authors and/or the copyright holders. The express permission of the copyright holder must be obtained for any use of this material other than for purposes permitted by law. •Users may freely distribute the URL that is used to identify this publication. •Users may download and/or print one copy of the publication from the University of Birmingham research portal for the purpose of private study or non-commercial research. •User may use extracts from the document in line with the concept of ‘fair dealing’ under the Copyright, Designs and Patents Act 1988 (?) •Users may not further distribute the material nor use it for the purposes of commercial gain. Where a licence is displayed above, please note the terms and conditions of the licence govern your use of this document. -

Astrocytoma: a Hormone-Sensitive Tumor?
International Journal of Molecular Sciences Review Astrocytoma: A Hormone-Sensitive Tumor? Alex Hirtz 1, Fabien Rech 1,2,Hélène Dubois-Pot-Schneider 1 and Hélène Dumond 1,* 1 Université de Lorraine, CNRS, CRAN, F-54000 Nancy, France; [email protected] (A.H.); [email protected] (F.R.); [email protected] (H.D.-P.-S.) 2 Université de Lorraine, CHRU-Nancy, Service de Neurochirurgie, F-54000 Nancy, France * Correspondence: [email protected]; Tel.: +33-372746115 Received: 29 October 2020; Accepted: 27 November 2020; Published: 30 November 2020 Abstract: Astrocytomas and, in particular, their most severe form, glioblastoma, are the most aggressive primary brain tumors and those with the poorest vital prognosis. Standard treatment only slightly improves patient survival. Therefore, new therapies are needed. Very few risk factors have been clearly identified but many epidemiological studies have reported a higher incidence in men than women with a sex ratio of 1:4. Based on these observations, it has been proposed that the neurosteroids and especially the estrogens found in higher concentrations in women’s brains could, in part, explain this difference. Estrogens can bind to nuclear or membrane receptors and potentially stimulate many different interconnected signaling pathways. The study of these receptors is even more complex since many isoforms are produced from each estrogen receptor encoding gene through alternative promoter usage or splicing, with each of them potentially having a specific role in the cell. The purpose of this review is to discuss recent data supporting the involvement of steroids during gliomagenesis and to focus on the potential neuroprotective role as well as the mechanisms of action of estrogens in gliomas. -

The New WHO Classification of Brain Tumors and Molecular Profiling in the Diagnosis of Gliomas
The New WHO Classification of Brain Tumors and Molecular Profiling in the Diagnosis of Gliomas Aivi Nguyen, MD Neuropathology Fellow Division of Neuropathology Center for Personalized Diagnosis (CPD) Glial neoplasms – infiltrating gliomas Astrocytic tumors • Diffuse astrocytoma II • Anaplastic astrocytoma III • Glioblastoma • Giant cell glioblastoma IV • Gliosarcoma Oligodendroglial tumors • Oligodendroglioma II • Anaplastic oligodendroglioma III Oligoastrocytic tumors • Oligoastrocytoma II • Anaplastic oligoastrocytoma III Courtesy of Dr. Maria Martinez-Lage 2 2016 3 The 2016 WHO classification of tumours of the central nervous system Louis et al., Acta Neuropathologica 2016 4 Talk Outline Genetic, epigenetic and metabolic changes in gliomas • Mechanisms/tumor biology • Incorporation into daily practice and WHO classification Penn’s Center for Personalized Diagnostics • Tests performed • Results and observations to date Summary 5 The 2016 WHO classification of tumours of the central nervous system Louis et al., Acta Neuropathologica 2016 6 Mechanism of concurrent 1p and 19q chromosome loss in oligodendroglioma lost FUBP1 CIC Whole-arm translocation Griffin et al., Journal of Neuropathology and Experimental Neurology 2006 7 Oligodendroglioma: 1p19q co-deletion Since the 1990s Diagnostic Prognostic Predictive Li et al., Int J Clin Exp Pathol 2014 8 Mutations of Selected Genes in Glioma Subtypes GBM Astrocytoma Oligodendroglioma Oligoastrocytoma Killela et al., PNAS 2013 9 Escaping Senescence Telomerase reverse transcriptase gene -

Malignant CNS Solid Tumor Rules
Malignant CNS and Peripheral Nerves Equivalent Terms and Definitions C470-C479, C700, C701, C709, C710-C719, C720-C725, C728, C729, C751-C753 (Excludes lymphoma and leukemia M9590 – M9992 and Kaposi sarcoma M9140) Introduction Note 1: This section includes the following primary sites: Peripheral nerves C470-C479; cerebral meninges C700; spinal meninges C701; meninges NOS C709; brain C710-C719; spinal cord C720; cauda equina C721; olfactory nerve C722; optic nerve C723; acoustic nerve C724; cranial nerve NOS C725; overlapping lesion of brain and central nervous system C728; nervous system NOS C729; pituitary gland C751; craniopharyngeal duct C752; pineal gland C753. Note 2: Non-malignant intracranial and CNS tumors have a separate set of rules. Note 3: 2007 MPH Rules and 2018 Solid Tumor Rules are used based on date of diagnosis. • Tumors diagnosed 01/01/2007 through 12/31/2017: Use 2007 MPH Rules • Tumors diagnosed 01/01/2018 and later: Use 2018 Solid Tumor Rules • The original tumor diagnosed before 1/1/2018 and a subsequent tumor diagnosed 1/1/2018 or later in the same primary site: Use the 2018 Solid Tumor Rules. Note 4: There must be a histologic, cytologic, radiographic, or clinical diagnosis of a malignant neoplasm /3. Note 5: Tumors from a number of primary sites metastasize to the brain. Do not use these rules for tumors described as metastases; report metastatic tumors using the rules for that primary site. Note 6: Pilocytic astrocytoma/juvenile pilocytic astrocytoma is reportable in North America as a malignant neoplasm 9421/3. • See the Non-malignant CNS Rules when the primary site is optic nerve and the diagnosis is either optic glioma or pilocytic astrocytoma.