Amidohydrolases of the Reductive Pyrimidine Catabolic Pathway Purification, Characterization, Structure, Reaction Mechanism
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Regulation of Carbamoyl Phosphate Synthetase-Aspartate Transcarbamoylase-Dihydroorotase (Cad) by Phosphorylation and Protein-Protein Interactions
THE REGULATION OF CARBAMOYL PHOSPHATE SYNTHETASE-ASPARTATE TRANSCARBAMOYLASE-DIHYDROOROTASE (CAD) BY PHOSPHORYLATION AND PROTEIN-PROTEIN INTERACTIONS Eric M. Wauson A dissertation submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Pharmacology. Chapel Hill 2007 Approved by: Lee M. Graves, Ph.D. T. Kendall Harden, Ph.D. Gary L. Johnson, Ph.D. Aziz Sancar M.D., Ph.D. Beverly S. Mitchell, M.D. 2007 Eric M. Wauson ALL RIGHTS RESERVED ii ABSTRACT Eric M. Wauson: The Regulation of Carbamoyl Phosphate Synthetase-Aspartate Transcarbamoylase-Dihydroorotase (CAD) by Phosphorylation and Protein-Protein Interactions (Under the direction of Lee M. Graves, Ph.D.) Pyrimidines have many important roles in cellular physiology, as they are used in the formation of DNA, RNA, phospholipids, and pyrimidine sugars. The first rate- limiting step in the de novo pyrimidine synthesis pathway is catalyzed by the carbamoyl phosphate synthetase II (CPSase II) part of the multienzymatic complex Carbamoyl phosphate synthetase, Aspartate transcarbamoylase, Dihydroorotase (CAD). CAD gene induction is highly correlated to cell proliferation. Additionally, CAD is allosterically inhibited or activated by uridine triphosphate (UTP) or phosphoribosyl pyrophosphate (PRPP), respectively. The phosphorylation of CAD by PKA and ERK has been reported to modulate the response of CAD to allosteric modulators. While there has been much speculation on the identity of CAD phosphorylation sites, no definitive identification of in vivo CAD phosphorylation sites has been performed. Therefore, we sought to determine the specific CAD residues phosphorylated by ERK and PKA in intact cells. -

Genome-Scale Fitness Profile of Caulobacter Crescentus Grown in Natural Freshwater
Supplemental Material Genome-scale fitness profile of Caulobacter crescentus grown in natural freshwater Kristy L. Hentchel, Leila M. Reyes Ruiz, Aretha Fiebig, Patrick D. Curtis, Maureen L. Coleman, Sean Crosson Tn5 and Tn-Himar: comparing gene essentiality and the effects of gene disruption on fitness across studies A previous analysis of a highly saturated Caulobacter Tn5 transposon library revealed a set of genes that are required for growth in complex PYE medium [1]; approximately 14% of genes in the genome were deemed essential. The total genome insertion coverage was lower in the Himar library described here than in the Tn5 dataset of Christen et al (2011), as Tn-Himar inserts specifically into TA dinucleotide sites (with 67% GC content, TA sites are relatively limited in the Caulobacter genome). Genes for which we failed to detect Tn-Himar insertions (Table S13) were largely consistent with essential genes reported by Christen et al [1], with exceptions likely due to differential coverage of Tn5 versus Tn-Himar mutagenesis and differences in metrics used to define essentiality. A comparison of the essential genes defined by Christen et al and by our Tn5-seq and Tn-Himar fitness studies is presented in Table S4. We have uncovered evidence for gene disruptions that both enhanced or reduced strain fitness in lake water and M2X relative to PYE. Such results are consistent for a number of genes across both the Tn5 and Tn-Himar datasets. Disruption of genes encoding three metabolic enzymes, a class C β-lactamase family protein (CCNA_00255), transaldolase (CCNA_03729), and methylcrotonyl-CoA carboxylase (CCNA_02250), enhanced Caulobacter fitness in Lake Michigan water relative to PYE using both Tn5 and Tn-Himar approaches (Table S7). -

Supplementary Materials
Supplementary Materials COMPARATIVE ANALYSIS OF THE TRANSCRIPTOME, PROTEOME AND miRNA PROFILE OF KUPFFER CELLS AND MONOCYTES Andrey Elchaninov1,3*, Anastasiya Lokhonina1,3, Maria Nikitina2, Polina Vishnyakova1,3, Andrey Makarov1, Irina Arutyunyan1, Anastasiya Poltavets1, Evgeniya Kananykhina2, Sergey Kovalchuk4, Evgeny Karpulevich5,6, Galina Bolshakova2, Gennady Sukhikh1, Timur Fatkhudinov2,3 1 Laboratory of Regenerative Medicine, National Medical Research Center for Obstetrics, Gynecology and Perinatology Named after Academician V.I. Kulakov of Ministry of Healthcare of Russian Federation, Moscow, Russia 2 Laboratory of Growth and Development, Scientific Research Institute of Human Morphology, Moscow, Russia 3 Histology Department, Medical Institute, Peoples' Friendship University of Russia, Moscow, Russia 4 Laboratory of Bioinformatic methods for Combinatorial Chemistry and Biology, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry of the Russian Academy of Sciences, Moscow, Russia 5 Information Systems Department, Ivannikov Institute for System Programming of the Russian Academy of Sciences, Moscow, Russia 6 Genome Engineering Laboratory, Moscow Institute of Physics and Technology, Dolgoprudny, Moscow Region, Russia Figure S1. Flow cytometry analysis of unsorted blood sample. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S2. Flow cytometry analysis of unsorted liver stromal cells. Representative forward, side scattering and histogram are shown. The proportions of negative cells were determined in relation to the isotype controls. The percentages of positive cells are indicated. The blue curve corresponds to the isotype control. Figure S3. MiRNAs expression analysis in monocytes and Kupffer cells. Full-length of heatmaps are presented. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Transcriptomic and Proteomic Profiling Provides Insight Into
BASIC RESEARCH www.jasn.org Transcriptomic and Proteomic Profiling Provides Insight into Mesangial Cell Function in IgA Nephropathy † † ‡ Peidi Liu,* Emelie Lassén,* Viji Nair, Celine C. Berthier, Miyuki Suguro, Carina Sihlbom,§ † | † Matthias Kretzler, Christer Betsholtz, ¶ Börje Haraldsson,* Wenjun Ju, Kerstin Ebefors,* and Jenny Nyström* *Department of Physiology, Institute of Neuroscience and Physiology, §Proteomics Core Facility at University of Gothenburg, University of Gothenburg, Gothenburg, Sweden; †Division of Nephrology, Department of Internal Medicine and Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, Michigan; ‡Division of Molecular Medicine, Aichi Cancer Center Research Institute, Nagoya, Japan; |Department of Immunology, Genetics and Pathology, Uppsala University, Uppsala, Sweden; and ¶Integrated Cardio Metabolic Centre, Karolinska Institutet Novum, Huddinge, Sweden ABSTRACT IgA nephropathy (IgAN), the most common GN worldwide, is characterized by circulating galactose-deficient IgA (gd-IgA) that forms immune complexes. The immune complexes are deposited in the glomerular mesangium, leading to inflammation and loss of renal function, but the complete pathophysiology of the disease is not understood. Using an integrated global transcriptomic and proteomic profiling approach, we investigated the role of the mesangium in the onset and progression of IgAN. Global gene expression was investigated by microarray analysis of the glomerular compartment of renal biopsy specimens from patients with IgAN (n=19) and controls (n=22). Using curated glomerular cell type–specific genes from the published literature, we found differential expression of a much higher percentage of mesangial cell–positive standard genes than podocyte-positive standard genes in IgAN. Principal coordinate analysis of expression data revealed clear separation of patient and control samples on the basis of mesangial but not podocyte cell–positive standard genes. -

Nucleotide Metabolism Pathway: the Achilles' Heel for Bacterial Pathogens
REVIEW ARTICLES Nucleotide metabolism pathway: the achilles’ heel for bacterial pathogens Sujata Kumari1,2,* and Prajna Tripathi1,3 1National Institute of Immunology, New Delhi 110 067, India 2Present address: Department of Zoology, Magadh Mahila College, Patna University, Patna 800 001, India 3Present address: Institute of Molecular Medicine, Jamia Hamdard, New Delhi 110 062, India de novo pathway, the nucleotides are synthesized from Pathogens exploit their host to extract nutrients for their survival. They occupy a diverse range of host simple precursor molecules. In the salvage pathway, the niches during infection which offer variable nutrients preformed nucleobases or nucleosides which are present accessibility. To cause a successful infection a patho- in the cell or transported from external environmental gen must be able to acquire these nutrients from the milieu to the cell are utilized to form nucleotides. host as well as be able to synthesize the nutrients on its own, if required. Nucleotides are the essential me- tabolite for a pathogen and also affect the pathophysi- Purine biosynthesis pathway ology of infection. This article focuses on the role of nucleotide metabolism of pathogens during infection The purine biosynthesis pathway is universally conserved in a host. Nucleotide metabolism and disease pathoge- in living organisms (Figure 1). As an example, we here nesis are closely related in various pathogens. Nucleo- present the pathway derived from well-studied Gram- tides, purines and pyrimidines, are biosynthesized by positive bacteria Lactococcus lactis. In the de novo the de novo and salvage pathways. Whether the patho- pathway the purine nucleotides are synthesized from sim- gen will employ the de novo or salvage pathway dur- ple molecules such as phosphoribosyl pyrophosphate ing infection is dependent on various factors, like (PRPP), amino acids, CO2 and NH3 by a series of enzy- availability of nucleotides, energy condition and pres- matic reactions. -

Mammalian Dihydroorotase: Nucleotide Sequence, Peptide
Proc. Natl. Acad. Sci. USA Vol. 87, pp. 174-178, January 1990 Biochemistry Mammalian dihydroorotase: Nucleotide sequence, peptide sequences, and evolution of the dihydroorotase domain of the multifunctional protein CAD (aspartate transcarbamylase/expression/pyrimidine biosynthesis/protein domains) JAMES P. SIMMER, RUTH E. KELLY*, AUSTIN G. RINKER, JR., BARBARA H. ZIMMERMANN, JOSHUA L. SCULLY, HYESOOK KIM, AND DAVID R. EVANS Department of Biochemistry, Wayne State University, Detroit, MI 48201 Communicated by Mary Ellen Jones, August 24, 1989 (received for review July 5, 1989) ABSTRACT Mammalian DHOase (S-dihydroorotate ami- Saul ull Ps, Puli P'jII KI)Er dohydrolase, EC 3.5.2.3) is part of a large multifunctional protein called CAD, which also has a carbamoyl-phosphate 3 8 40% 4 44 4 synthetase [carbon-dioxide:L-glutamine amido-ligase (ADP- forming, carbamate-phosphorylating), EC 6.3.5.5] and aspar- tate transcarbamoylase (carbamoyl-phosphate:L-aspartate carbamoyltransferase, EC 2.1.3.2) activities. We sequenced FIG. 1. Nucleotide sequencing strategy. The region of the selected restriction fragments of a Syrian hamster CAD cDNA. pCAD142 sequenced is shown schematically: the CPSase domain The deduced amino acid sequence agreed with the sequence of (stippled bar), the DHOase domain (shaded bar), and the DHOase- tryptic peptides and the amino acid composition ofthe DHOase ATCase linker (clear bar). Map units represent the distance in domain isolated by controlled proteolysis of CAD. Escherichia kilobases from the start of the cDNA insert. Clones sequenced are coli transformed with a recombinant plasmid containing the indicated by arrows. cDNA segment 5' to the aspartate transcarbamoylase coding respectively, may participate in catalysis. -

Using Peptides to Examine the Interaction Interface Between Aspartate Transcarbamoylase and Dihydroorotase in Pyrimidine Biosynthesis in Aquifex Aeolicus Nouf Alyami
Eastern Michigan University DigitalCommons@EMU Master's Theses, and Doctoral Dissertations, and Master's Theses and Doctoral Dissertations Graduate Capstone Projects 7-14-2015 Using peptides to examine the interaction interface between Aspartate transcarbamoylase and Dihydroorotase in pyrimidine biosynthesis in Aquifex aeolicus Nouf Alyami Follow this and additional works at: http://commons.emich.edu/theses Part of the Chemistry Commons Recommended Citation Alyami, Nouf, "Using peptides to examine the interaction interface between Aspartate transcarbamoylase and Dihydroorotase in pyrimidine biosynthesis in Aquifex aeolicus" (2015). Master's Theses and Doctoral Dissertations. 641. http://commons.emich.edu/theses/641 This Open Access Thesis is brought to you for free and open access by the Master's Theses, and Doctoral Dissertations, and Graduate Capstone Projects at DigitalCommons@EMU. It has been accepted for inclusion in Master's Theses and Doctoral Dissertations by an authorized administrator of DigitalCommons@EMU. For more information, please contact [email protected]. Using Peptides to Examine the Interaction Interface between Aspartate transcarbamoylase and Dihydroorotase in Pyrimidine Biosynthesis in Aquifex aeolicus by Nouf Alyami Thesis Submitted to the Department of Chemistry Eastern Michigan University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE in Chemistry Thesis Committee: Hedeel Evans, Ph.D., Chair Deborah Heyl-Clegg, Ph.D. Vance Kennedy, Ph.D. July 14, 2015 Ypsilanti, Michigan ii DEDICATION This work is dedicated to my parents, without whose support it would not have been possible. A special feeling of gratitude to my loving siblings Nada, Bayan, and Bader whose laughter is like music to my soul. Also, I dedicate this work to my two grandmothers whose hearts are full of prayers for me. -

Dihydropyrimidine Dehydrogenase in the Metabolism of the Anticancer Drugs
Cancer Chemotherapy and Pharmacology (2019) 84:1157–1166 https://doi.org/10.1007/s00280-019-03936-w REVIEW ARTICLE Dihydropyrimidine dehydrogenase in the metabolism of the anticancer drugs Vinay Sharma1 · Sonu Kumar Gupta1 · Malkhey Verma1 Received: 3 May 2019 / Accepted: 21 August 2019 / Published online: 4 September 2019 © Springer-Verlag GmbH Germany, part of Springer Nature 2019 Abstract Cancer caused by fundamental defects in cell cycle regulation leads to uncontrolled growth of cells. In spite of the treatment with chemotherapeutic agents of varying nature, multiple resistance mechanisms are identifed in cancer cells. Similarly, numerous variations, which decrease the metabolism of chemotherapeutics agents and thereby increasing the toxicity of anticancer drugs have been identifed. 5-Fluorouracil (5-FU) is an anticancer drug widely used to treat many cancers in the human body. Its broad targeting range is based upon its capacity to act as a uracil analogue, thereby disrupting RNA and DNA synthesis. Dihydropyrimidine dehydrogenase (DPD) is an enzyme majorly involved in the metabolism of pyrimidines in the human body and has the same metabolising efect on 5-FU, a pyrimidine analogue. Multiple mutations in the DPD gene have been linked to 5-FU toxicity and inadequate dosages. DPD inhibitors have also been used to inhibit excessive degradation of 5-FU for meeting appropriate dosage requirements. This article focusses on the role of dihydropyrimidine dehydrogenase in the metabolism of the anticancer drug 5-FU and other associated drugs. Keywords Cancer · Anticancer drugs · Dihydropyrimidine dehydrogenase (DPD) · 5-Fluorouracil (5-FU) · Drug resistance · Drug metabolism Introduction by Dihydropyrimidine dehydrogenase (DPD) through the pyrimidine degradation pathway. -

PURINE SALVAGE in HELICOBACTER PYLORI by ERICA FRANCESCA MILLER (Under the Direction of Robert J. Maier) ABSTRACT Purines Are Es
PURINE SALVAGE IN HELICOBACTER PYLORI by ERICA FRANCESCA MILLER (Under the Direction of Robert J. Maier) ABSTRACT Purines are essential for all living cells. This fact is reflected in the high degree of pathway conservation for purine metabolism across all domains of life. The availability of purines within a mammalian host is thought to be a limiting factor for infection, as demonstrated by the importance of purine synthesis and salvage genes among many bacterial pathogens. Helicobacter pylori, a primary causative agent of peptic ulcers and gastric cancers, colonizes a niche that is otherwise uninhabited by bacteria: the surface of the human gastric epithelium. Despite many studies over the past 30 years that have addressed virulence mechanisms such as acid resistance, little knowledge exists regarding this organism’s purine metabolism. To fill this gap in knowledge, we asked whether H. pylori can carry out de novo purine biosynthesis, and whether its purine salvage network is complete. Based on genomic data from the fully sequenced H. pylori genomes, we combined mutant analysis with physiological studies to determine that H. pylori, by necessity, must acquire purines from its human host. Furthermore, we found the purine salvage network to be complete, allowing this organism to use any single purine nucleobase or nucleoside for growth. In the process of elucidating these pathways, we discovered a nucleoside transporter in H. pylori that, in contrast to the biochemically- characterized homolog NupC, aids in uptake of purine rather than pyrimidine nucleosides into the cell. Lastly, we investigated an apparent pathway gap in the genome annotation—that of adenine degradation—and in doing so uncovered a new family of adenosine deaminase that lacks sequence homology with all other adenosine deaminases studied to date. -
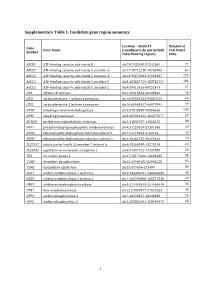
Supplementary Table 1. Candidate Gene Region Summary
Supplementary Table 1. Candidate gene region summary Location – Build 37 Number of Gene Gene Name (coordinates do not include Test Panel Symbol 25kb flanking region) SNPs ABCB1 ATP-binding cassette, sub-family B chr7:87132948-87342564 77 ABCC3 ATP-binding cassette, sub-family C, member 3 chr17:48712218-48769063 64 ABCC4 ATP-binding cassette, sub-family C, member 4 chr13:95672083-95953687 224 ABCC5 ATP-binding cassette, sub-family C, member 5 chr3:183637724-183735727 101 ABCG2 ATP-binding cassette, sub-family G, member 2 chr4:89011416-89152474 57 CDA cytidine deaminase chr1:20915444-20945400 26 CES1 carboxylesterase 1 isoform a precursor chr16:55836764-55867075 24 CES2 carboxylesterase 2 isoform a precursor chr16:66968347-66978994 59 DPYD dihydropyrimidine dehydrogenase chr1:97543300-98386615 239 DPYS dihydropyrimidinase chr8:105391652-105479277 69 MTHFR methylenetetrahydrofolate reductase chr1:11845787-11866115 38 PPAT phosphoribosyl pyrophosphate amidotransferase chr4:57259529-57301845 29 RRM1 ribonucleoside-diphosphate reductase subunit 1 chr11:4115924-4160106 29 RRM2 ribonucleoside-diphosphate reductase subunit 2 chr2:10262735-10270623 19 SLC22A7 solute carrier family 22 member 7 isoform b chr6:43265998-43273276 26 SLC29A1 equilibrative nucleoside transporter 1 chr6:44187242-44201888 26 TK1 thymidine kinase 1 chr17:76170160-76183285 35 TYMP thymidine phosphorylase chr22:50964182-50968258 92 TYMS thymidylate synthetase chr18:657604-673499 34 UCK1 uridine-cytidine kinase 1 isoform a chr9:134399191-134406655 43 UCK2 uridine-cytidine kinase 2 isoform a chr1:165796890-165877339 22 UMPS uridine monophosphate synthase chr3:124449213-124464040 34 UPB1 beta-ureidopropionase chr22:24890077-24922553 30 UPP1 uridine phosphorylase 1 chr7:48128355-48148330 16 UPP2 uridine phosphorylase 2 chr2:158851691-158992478 43 1 Supplementary Table 2. -

The De Novo Biosynthesis of Uridine Monophosphate (UMP) Involves Six Enzymatic Reactions and Appears to Be Encoded by Only Three Structural Genes in Animals
J. Nutr. Sci. Vitaminol., 37, 517-528, 1991 Effect of Dietary Protein on Pyrimidine-Metabolizing Enzymes in Rats Masae KANEKO, Shigeko FUJIMOTO, Mariko KIKUGAWA, Yasuhide KONTANI, and Nanaya TAMAKI* Laboratory of Nutritional Chemistry, Faculty of Nutrition, Kobe-Gakuin University, Nishi-ku, Kobe 651-21, Japan (Received February 25, 1991) Summary The effect of dietary protein on pyrimidine-metabolizing enzymes was studied in the rat. The activities of dihydropyrimidine dehydrogenase and ƒÀ-ureidopropionase in the livers of rats fed a protein free diet were significantly decreased, while the activity of dihydropyrim idinase was unaffected. Protein deficiency (5%) also decreased the activity of ƒÀ-ureidopropionase. On the other hand, a high-protein diet (60%) increased the level of ƒÀ-ureidopropionase. The activities of ƒÀ- alanine-oxoglutarate aminotransferase (aminobutyrate aminotransferase) and D-3-aminoisobutyrate-pyruvate aminotransferase ((R)-3-amino-2- methylpropionate-pyruvate aminotransferase), which are present in mi tochondria, depended on the amount of protein in the diet. Ammonium ions supplemented in the diet and given by injection did not affect the activities of rat liver pyrimidine-metabolizing enzymes (dihydropyrimi dine dehydrogenase, dihydropyrimidinase, ƒÀ-ureidopropionase, ƒÀ-alanine - oxoglutarate aminotransferase and D-3-aminoisobutyrate-pyruvate ami notransferase). Dietary uridine resulted in the accumulation of uracil in the liver, but did not affect the activities of pyrimidine-metabolizing enzymes. Key Words dihydropyrimidine dehydrogenase, dihydropyrimidinase, ƒÀ- ureidopropionase, ƒÀ-alanine-oxoglutarate aminotransferase, D-3-amino isobutyrate-pyruvate aminotransferase, pyrimidine The de novo biosynthesis of uridine monophosphate (UMP) involves six enzymatic reactions and appears to be encoded by only three structural genes in animals. The active sites of the first three enzymes, carbamoyl-phosphate synthase, aspartate transcarbamylase, and dihydroorotase, are on a single large polypeptide * To whom correspondence should be addressed .