Antimicrobial Use and Resistance (AUR) Module
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Piperacillin-Tazobactam Allergies: an Exception to Usual Penicillin Allergy
Allergy Asthma Immunol Res. 2021 Mar;13(2):284-294 https://doi.org/10.4168/aair.2021.13.2.284 pISSN 2092-7355·eISSN 2092-7363 Original Article Piperacillin-Tazobactam Allergies: An Exception to Usual Penicillin Allergy Jane CY Wong ,1 Elaine YL Au ,2 Heather HF Yeung ,2 Chak-Sing Lau ,1 Philip Hei Li 1* 1Division of Rheumatology and Clinical Immunology, Department of Medicine, The University of Hong Kong, Queen Mary Hospital, Hong Kong 2Division of Clinical Immunology, Department of Pathology, The University of Hong Kong, Queen Mary Hospital, Hong Kong Received: Apr 21, 2020 Revised: Jul 20, 2020 ABSTRACT Accepted: Jul 26, 2020 Purpose: The majority of penicillin allergy labels are false, and skin tests (ST) have high Correspondence to negative predictive value (NPV) of up to 90%. Piperacillin-tazobactam (PT) allergy has been Philip Hei Li, MBBS, FHKCP suspected to be an exception to this, but existing literature is scarce. We investigate the Division of Rheumatology & Clinical Immunology, Department of Medicine, The epidemiology, clinical characteristics, testing outcomes and predictive value of ST in patients University of Hong Kong, Queen Mary Hospital, referred for suspected PT allergies. 102 Pokfulam Road, Hong Kong. Methods: The records of all patients referred for suspected PT allergy testing and Tel: +852-2255-3348 prescription rates of PT in all Hong Kong public hospitals (2015–2019) were analyzed. Fax: +852-2816-2863 Results: There was an increase in both PT prescriptions and number of newly reported E-mail: [email protected] PT allergies between 2015 and 2019. The majority (91.1%) of patients with suspected PT Copyright © 2021 The Korean Academy of allergy had at least 1 underlying medical co-morbidity or immunosuppressant use leading to Asthma, Allergy and Clinical Immunology • increased risk of infections. -

Anew Drug Design Strategy in the Liht of Molecular Hybridization Concept
www.ijcrt.org © 2020 IJCRT | Volume 8, Issue 12 December 2020 | ISSN: 2320-2882 “Drug Design strategy and chemical process maximization in the light of Molecular Hybridization Concept.” Subhasis Basu, Ph D Registration No: VB 1198 of 2018-2019. Department Of Chemistry, Visva-Bharati University A Draft Thesis is submitted for the partial fulfilment of PhD in Chemistry Thesis/Degree proceeding. DECLARATION I Certify that a. The Work contained in this thesis is original and has been done by me under the guidance of my supervisor. b. The work has not been submitted to any other Institute for any degree or diploma. c. I have followed the guidelines provided by the Institute in preparing the thesis. d. I have conformed to the norms and guidelines given in the Ethical Code of Conduct of the Institute. e. Whenever I have used materials (data, theoretical analysis, figures and text) from other sources, I have given due credit to them by citing them in the text of the thesis and giving their details in the references. Further, I have taken permission from the copyright owners of the sources, whenever necessary. IJCRT2012039 International Journal of Creative Research Thoughts (IJCRT) www.ijcrt.org 284 www.ijcrt.org © 2020 IJCRT | Volume 8, Issue 12 December 2020 | ISSN: 2320-2882 f. Whenever I have quoted written materials from other sources I have put them under quotation marks and given due credit to the sources by citing them and giving required details in the references. (Subhasis Basu) ACKNOWLEDGEMENT This preface is to extend an appreciation to all those individuals who with their generous co- operation guided us in every aspect to make this design and drawing successful. -

Antimicrobial Resistance of Bacteraemia in the Emergency
Rothe et al. BMC Infectious Diseases (2019) 19:1091 https://doi.org/10.1186/s12879-019-4721-9 RESEARCH ARTICLE Open Access Antimicrobial resistance of bacteraemia in the emergency department of a German university hospital (2013–2018): potential carbapenem-sparing empiric treatment options in light of the new EUCAST recommendations Kathrin Rothe1,2* , Nina Wantia1,2, Christoph D. Spinner2,3, Jochen Schneider2,3, Tobias Lahmer2,3, Birgit Waschulzik4, Roland M. Schmid2,3, Dirk H. Busch1,2 and Juri Katchanov2,3 Abstract Background: This study investigated predominant microorganisms causing community-onset bacteraemia at the medical emergency department (ED) of a tertiary-care university hospital in Germany from 2013 to 2018 and their antimicrobial susceptibility patterns. Methods: Antimicrobial resistance patterns in patients with positive blood cultures presenting to an internal medicine ED were retrospectively analysed. Results: Blood cultures were obtained at 5191 of 66,879 ED encounters, with 1013 (19.5%) positive results, and true positive results at 740 encounters (diagnostic yield, 14.3%). The most frequently isolated relevant microorganisms were Enterobacterales (n = 439, 59.3%), Staphylococcus aureus (n = 92, 12.4%), Streptococcus pneumoniae (n = 34, 4.6%), Pseudomonas aeruginosa (n = 32, 4.3%), Streptococcus pyogenes (n = 16, 2.2%), Enterococcus faecalis (n = 18, 2.4%), and Enterococcus faecium (n = 12, 1.6%). Antimicrobial susceptibility testing revealed a high proportion of resistance against ampicillin-sulbactam in Enterobacterales (42.2%). The rate of methicillin-resistant Staphylococcus aureus was low (0.4%). Piperacillin-tazobactam therapy provided coverage for 83.2% of all relevant pathogens using conventional breakpoints. Application of the new European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendations increased the percentage of susceptible isolates to high-dose piperacillin-tazobactam to 92.8% (p < 0.001). -

BMJ Open Is Committed to Open Peer Review. As Part of This Commitment We Make the Peer Review History of Every Article We Publish Publicly Available
BMJ Open: first published as 10.1136/bmjopen-2018-027935 on 5 May 2019. Downloaded from BMJ Open is committed to open peer review. As part of this commitment we make the peer review history of every article we publish publicly available. When an article is published we post the peer reviewers’ comments and the authors’ responses online. We also post the versions of the paper that were used during peer review. These are the versions that the peer review comments apply to. The versions of the paper that follow are the versions that were submitted during the peer review process. They are not the versions of record or the final published versions. They should not be cited or distributed as the published version of this manuscript. BMJ Open is an open access journal and the full, final, typeset and author-corrected version of record of the manuscript is available on our site with no access controls, subscription charges or pay-per-view fees (http://bmjopen.bmj.com). If you have any questions on BMJ Open’s open peer review process please email [email protected] http://bmjopen.bmj.com/ on September 26, 2021 by guest. Protected copyright. BMJ Open BMJ Open: first published as 10.1136/bmjopen-2018-027935 on 5 May 2019. Downloaded from Treatment of stable chronic obstructive pulmonary disease: a protocol for a systematic review and evidence map Journal: BMJ Open ManuscriptFor ID peerbmjopen-2018-027935 review only Article Type: Protocol Date Submitted by the 15-Nov-2018 Author: Complete List of Authors: Dobler, Claudia; Mayo Clinic, Evidence-Based Practice Center, Robert D. -

Β-Lactam Overview Along with Mezlocillin and Azlocillin
Piperacillin* Class: β-lactam Overview Along with mezlocillin and azlocillin, piperacillin is a ureidopenicillin developed for additional activity against Klebsiella and Enterobacter species, and Pseudomonas aeruginosa. The drug is a derivative of ampicillin, an acyl side chain along with a piperazine group is added to the original molecule, and has similar activity against streptococcal species. Piperacillin is poorly absorbed because it is hydrolyzed by acids in the stomach. Carboxypenicillins, another pencillin group with enhanced activity against Pseudomonas aeruginosa and ureidopenicillins can precipitate the side effects of hypokalemia and hypernatremia, although side effects are generally less common with the latter. Piperacillin also acts through interference with cell wall synthesis. Resistance Some Pseudomonas aeruginosa strains have developed resistance to piperacillin by plasmid or chromosomal transfer of the ability to produce effective β-lactamases. Effectiveness Piperacillin and, in general, the ureidopenicillins are utilized primarily against Pseudomonas infections of the urinary tract, lung and blood. The drug also is very effective against Enterococcus species. In addition the ureidopenicillins exhibit enhanced activity against other aerobic Gram-negative organisms. Piperacillin specifically is used in infections caused by several Shigella and Proteus species and a few Citrobacter and Enterobacter species. This antimicrobial is also commonly used in the treatment of burn patients. Piperacillin is often combined with tazobactam, a β-lactamase inhibitor, to combat the production of β-lactamases by Gram-negative bacteria. *References available by request. Call the Infectious Disease Epidemiology Section, Office of Public Health, Louisiana Department of Health and Hospitals (504-219-4563) . -

Guidelines on Urological Infections
Guidelines on Urological Infections M. Grabe (Chairman), T.E. Bjerklund-Johansen, H. Botto, M. Çek, K.G. Naber, P. Tenke, F. Wagenlehner © European Association of Urology 2010 TABLE OF CONTENTS PAGE 1. INTRODUCTION 7 1.1 Pathogenesis of urinary tract infections 7 1.2 Microbiological and other laboratory findings 7 1.3 Classification of urological infections 8 1.4 Aim of guidelines 8 1.5 Methods 9 1.6 Level of evidence and grade of guideline recommendations 9 1.7 References 9 2. UNCOMPLICATED URINARY TRACT INFECTIONS IN ADULTS 11 2.1 Definition 11 2.1.1 Aetiological spectrum 11 2.2 Acute uncomplicated cystitis in premenopausal, non-pregnant women 11 2.2.1 Diagnosis 11 2.2.1.1 Clinical diagnosis 11 2.2.1.2 Laboratory diagnosis 11 2.2.2 Therapy 11 2.2.3 Follow up 12 2.3 Acute uncomplicated pyelonephritis in premenopausal, non-pregnant women 12 2.3.1 Diagnosis 12 2.3.1.1 Clinical diagnosis 12 2.3.1.2 Laboratory diagnosis 12 2.3.1.3 Imaging diagnosis 13 2.3.2 Therapy 13 2.3.2.1 Mild and moderate cases of acute uncomplicated pyelonephritis 13 2.3.2.2 Severe cases of acute uncomplicated pyelonephritis 13 2.3.3 Follow-up 14 2.4 Recurrent (uncomplicated) UTIs in women 16 2.4.1 Diagnosis 16 2.4.2 Prevention 16 2.4.2.1 Antimicrobial prophylaxis 16 2.4.2.2 Immunoactive prophylaxis 16 2.4.2.3 Prophylaxis with probiotics 17 2.4.2.4 Prophylaxis with cranberry 17 2.5 Urinary tract infections in pregnancy 17 2.5.1 Definition of significant bacteriuria 17 2.5.2 Screening 17 2.5.3 Treatment of asymptomatic bacteriuria 17 2.5.4 Duration of therapy 18 2.5.5 Follow-up 18 2.5.6 Prophylaxis 18 2.5.7 Treatment of pyelonephritis 18 2.5.8 Complicated UTI 18 2.6 UTIs in postmenopausal women 18 2.6.1 Risk factors 18 2.6.2 Diagnosis 18 2.6.3 Treatment 18 2.7 Acute uncomplicated UTIs in young men 19 2.7.1 Men with acute uncomplicated UTI 19 2.7.2 Men with UTI and concomitant prostate infection 19 2.8 Asymptomatic bacteriuria 19 2.8.1 Diagnosis 19 2.8.2 Screening 19 2.9 References 26 3. -

VITEK® 2 Susceptibility Cards for Gram Negative Bacilli
VITEK® 2 Susceptibility Cards for Gram Negative Bacilli GN66 GN67 GN68 GN69 GN70 GN71 GN72 GN73 GN74 GN75 GN76 GN77 GN79 GN80 GN81 GN82 GN83 GN84 GN86 GN87 GN90 GN91 GN92 GN93 *XN06 Class Antibiotic MIC Calling Range 413398 413399 413431 413400 413401 413402 413403 413404 413941 413432 413433 413434 413436 413437 413438 413439 413440 413410 413942 413943 414779 414780 414963 414985 413944 Amoxicillin / Clavulanic Acid 2/1 - 32/16 Aminopenicillin / inhibitor Ampicillin 2 - 32 combination Ampicillin / Sulbactam 2/1 - 32/16 Piperacillin 4 - 128 Piperacilllin / Tazobactam 4/4 - 128/4 Ureidopenicillin / inhibitor combinations Ticarcillin 8 - 128 Ticarcillin / Clavulanic 8/2 - 128/2 Acid Cefalotin 2 - 64 Cephalosporin I Cefazolin 4 - 64 Cefotetan 4 - 64 Cephalosporin II / Cefoxitin 4 - 64 Cephamycin Cefuroxime 1 - 64 Cefepime 1 - 64 Cefotaxime 1 - 64 Cefpodoxime 0.25 - 8 Cephalosporin III / IV Ceftazidime 1 - 64 Ceftizoxime 1 - 64 Ceftriaxone 1 - 64 Monobactam Aztreonam 1 - 64 ESBL ESBL Confirmation Test +/- Doripenem 0.12 - 8 Ertapenem 0.5 - 8 Carbapenem Imipenem 0.25 - 16 Meropenem 0.25 - 16 Amikacin 2 - 64 Aminoglycoside Gentamicin 1 - 16 Tobramycin 1 - 16 Ciprofloxacin 0.25 - 4 Levofloxacin 0.12 - 8 Fluroquinolone Moxifloxacin 0.25 - 8 Norfloxacin 16 - 512 Tetracycline 1 - 16 Tetracyclines Tigecycline 0.5 - 8 Nalidixic Acid 2 - 32 Nitrofurantoin 16 - 512 Miscellaneous Trimethoprim / 20 (1/19) - 320 Sulfamethoxazole (16/304) Go to www.totalast.com for more information and to get help in selecting your next VITEK 2® card. *mates to GN69 ©2018 BIOMÉRIEUX, INC. THE BLUE LOGO AND VITEK ARE REGISTERED TRADEMARKS OF BIOMÉRIEUX. PRINTED IN USA. 17 0285 02 Susceptibility Cards for Gram Positive Cocci AST-GP74 414971 S. -
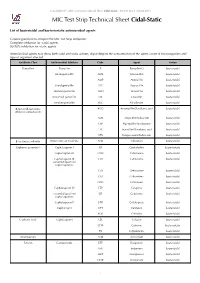
MIC Test Strip Technical Sheet Cidal-Static - MTS30 Rev.3 / 08.06.2015 MIC Test Strip Technical Sheet Cidal-Static
© Liofilchem® - MIC Test Strip Technical Sheet Cidal-Static - MTS30 Rev.3 / 08.06.2015 MIC Test Strip Technical Sheet Cidal-Static List of bactericidal and bacteriostatic antimicrobial agents General guideline to interpret the MIC Test Strip endpoints: Complete inhibition for -cidal agents. 80-90% inhibition for -static agents. Antimicrobial agents may show both cidal and static actions, depending on the concentration of the agent, count of microorganism and type of organism affected. Antibiotic Class Antimicrobial Subclass Code Agent Action Penicillins Penicillin P Penicillin G bactericidal Aminopenicillin AML Amoxicillin bactericidal AMP Ampicillin bactericidal Ureidopenicillin PIP Piperacillin bactericidal Methoxypenicillin TMO Temocillin bactericidal Isoxazolyl penicillin OX Oxacillin bactericidal Amidinopenicillin MEC Mecillinam bactericidal β-lactam/β-lactamase AUG Amoxicillin/Clavulanic acid bactericidal inhibitor combinations AMS Ampicillin/Sulbactam bactericidal TZP Piperacillin/Tazobactam bactericidal TTC Ticarcillin/Clavulanic acid bactericidal CPS Cefoperazone/Sulbactam bactericidal β-lactamase inhibitor Penicillanic acid sulfone ATM Sulbactam bactericidal Cephems (parenteral) Cephalosporin I KF Cephalothin bacteriostatic Cephalosporin II CXM Cefuroxime bactericidal Cephalosporin III CTX Cefotaxime bactericidal (extended spectrum cephalosporins) CZX Ceftizoxime bactericidal CAZ Ceftazidime bactericidal CRO Ceftriaxone bactericidal Cephalosporin IV FEP Cefepime bactericidal (extended spectrum CR Cefpirome bactericidal cephalosporins) -

Antibiotics in the Treatment of Biliary Infection
Gut: first published as 10.1136/gut.25.9.988 on 1 September 1984. Downloaded from Gut, 1984, 25, 988-998 Progress report Antibiotics in the treatment of biliary infection Bile is difficult to obtain for culture in man. Because of this, antibiotic treatment of biliary infection is often directed towards biliary organisms which are suspected, rather than proven. To complicate effective treatment further, infection may be asymptomatic, presenting with septic complications following a surgical or radiological procedure. Despite knowledge of the pattern of infection in biliary disease, the microorganisms involved, and the spectrum and pharmacokinetics of potentially effective antibiotics, bacteria are often difficult to eradicate from the bile and biliary surgery has a high incidence of septic complications. There have been few reviews of this subject, particularly incorporating recent data, and a reassessment is necessary. This paper describes the clinical and bacteriological aspects of biliary infection, and the excretion and efficacy of specific antibiotics in bile. Based on the information available a therapeutic strategy is proposed. Biliary infection http://gut.bmj.com/ DEFINITION Ideally biliary tract infection should be defined by the organism count in bile. Keighley et al' reported finding at least 105 organisms per ml in over 90% of positive peroperative bile cultures, and this count could be taken as diagnostic. Such a definition is, however, impracticable because bile is difficult to collect. Although biliary sepsis is diagnosed clinically when there are systemic signs of infection combined with features of biliary tract on September 27, 2021 by guest. Protected copyright. disease, organisms may be cultured from bile in the absence of symptoms. -

VITEK® 2 Susceptibility Cards for Gram-Negative Bacilli
VITEK® 2 Susceptibility Cards for Gram-negative Bacilli GN66 GN67 GN69 GN70 GN72 GN73 GN74 GN77 GN79 GN80 GN81 GN83 GN84 GN93 GN95 GN99 N800† N801 N802 XN09* XN15‡ Class Antibiotic MIC Calling Range 413398 413399 413400 413401 413403 413404 413941 413434 413436 413437 413438 413440 413410 414985 421982 423102 423310 423416 423706 423425 423829 Ampicillin 2 - 32 Aminopenicillin & Ampicillin / Sulbactam 2/1 - 32/16 inhibitor combination Amoxicillin / Clavulanic Acid 2/1 - 32/16 Ureidopenicillin / Piperacilllin / Tazobactam 4/4 - 128/4 inhibitor combinations Ticarcillin / Clavulanic Acid 8/2 - 128/2 Ceftazidme/Avibactam 0.12 - 16 -lactam / β Ceftolozane/Tazobactam 0.25 - 32 inhibitor combinations Meropenem /Vaborbactam Cefalotin 2 - 64 Cephalosporin I Cefazolin 4 - 64 Cefotetan 4 - 64 Cephalosporin II / Cefoxitin 4 - 64 Cephamycin Cefuroxime 1 - 64 Cefotaxime 1 - 64 Cefotaxime (reformulated) 0.25 - 64 Cefpodoxime 0.25 - 8 Ceftazidime 1 - 64 Cephalosporin III / IV Ceftriaxone 1 - 64 Ceftriaxone (reformulated) 0.25 - 64 Cefepime 1 - 64 Cefepime (reformulated) 0.12 - 32 Monobactam Aztreonam 1 - 64 ESBL ESBL Confirmation Test +/- Ertapenem 0.5 - 8 Ertapenem (reformulated) 0.12 - 8 Carbapenem Imipenem 0.25 - 16 Imipenem (reformulated) 0.25 - 16 Meropenem 0.25 - 16 Amikacin 2 - 64 Amikacin (reformulated) 1 - 64 Gentamicin 1 - 16 Aminoglycoside Gentamicin (reformulated) 1 - 16 Tobramycin 1 - 16 Tobramycin (reformulated) 1 - 16 Ciprofloxacin 0.25 - 4 Ciprofloxacin (reformulated) 0.06 - 4 Fluoroquinolone Levofloxacin 0.12 - 8 Moxifloxacin 0.25 - 8 Doxycycline 0.5 - 16 Minocycline 0.5 - 32 Tetracyclines Tetracycline 1 - 16 Tigecycline 0.5 - 8 Tigecycline (reformulated) 0.5 - 8 Nalidixic Acid 2 - 32 Miscellaneous Nitrofurantoin 16 - 512 Trimethoprim / Sulfamethoxazole 20 (1/19) - 320 (16/304) *mates to GN69 & GN95 ‡ mates to N802 †Note: Deduce ceftriaxone based on cefotaxime Susceptibility Cards for Gram-positive Cocci (Staphylococcus spp., Enterococcus spp. -

Myelosuppression in Patients with Prolonged Use of Piperacillin/Tazobactam
DOI: 10.14744/ejmi.2019.84589 EJMI 2020;4(1):7–11 Research Article Myelosuppression in Patients with Prolonged use of Piperacillin/Tazobactam Ahmet Riza Sahin,1 Ali Muhittin Tasdogan2 1Department of Infection Diseases and Clinical Microbiology, Sutcu Imam University Faculty of Medicine, Kahramanmaras, Turkey 2Department of Anesthesiology and Reanimation, Hasan Kalyoncu University Health Sciences Faculty, Gaziantep, Turkey Abstract Objectives: The use of piperacillin-tazobactam in hospital acquired infections requiring long treatment periods may cause adverse effects including myelosuppression. Myelosuppression results in rare, but potentially serious clinic mani- festations such as neutropenia, thrombocytopenia, leukemia and anemia. The objective of this study was to investigate the incidence and characteristics of myelosuppression in patients with prolonged use of piperacillin-tazobactam. Methods: Inpatients followed-up and treated in Kahramanmaras Sutcu Imam University Medical Faculty Hospital and MMT Gaziantep American Hospital between April 1, 2017 and December 31, 2018 were included in the study. Patients’ demographic data, biochemical laboratory outcome, duration and dose of antibiotic treatment, comorbidities, side effects of antibiotic therapy were recorded and analyzed. Results: A total of 34 inpatients who received antibiotic therapy with piperacillin-tazobactam due to various diagnoses were included in the study. The mean duration of PTZ use was found as 11.9±6.31 days. Of all patients, 19 (55.9%) used antibiotics for longer than 10 days, while 15 (44.1%) used PTZ for 10 days or shorter. The mean duration of antibiotic use was found as 12.2 days in patients aged 65 years and over, while this duration was 11.5 days in patients aged under 65 years. -
CARBAPENEM RESISTANCE from Diagnosis to Outbreak Management TABLE of CONTENTS INTRODUCTION
CARBAPENEM RESISTANCE From diagnosis to outbreak management TABLE OF CONTENTS INTRODUCTION CARBAPENEM RESISTANCE p. 2 Bacterial resistance to antibiotics has become a major public health 1 issue worldwide. The reality of this threat was acknowledged in the WHO 2014 report (www.who.int/drug resistance/en) on antibiotic 2 EPIDEMIOLOGY p. 6 resistance. Rising resistance is of particular concern for Gram-negative bacilli 3 CLINICAL ASPECTS p. 11 such as Pseudomonas aeruginosa, Acinetobacter baumannii and Enterobacteriaceae, the latter being the most important pathogens for mankind. Carbapenems are last resort antibiotics for treating 4 TREATMENT p. 12 infections due to these Gram-negative bacilli (31). Resistance to carbapenems in these species is related either to 5 DIAGNOSIS p. 14 combined mechanisms of resistance (overexpression of broad- spectrum ß-lactamases together with efflux pumps, impermeability) SCREENING p. 20 or expression of carbapenem-hydrolyzing ß-lactamases, known as 6 carbapenemases (31). INFECTION CONTROL AND In Enterobacteriaceae, carbapenemases represent the most important 7 PREVENTION p. 23 mechanism of resistance, since the carbapenemase genes are mostly plasmid-encoded, associated with multi- or pan-drug resistance and are highly transferable, at least within the enterobacterial species, making them potentially responsible for outbreaks (31, 36, 38). CONCLUSION p. 25 LIST OF ABBREVIATIONS p. 26 This booklet covers issues related to carbapenem-resistant Gram-negative bacilli (mostly carbapenemase producers in REFERENCES