Genetic Diversity in Kiwifruit Polyploid Complexes: Insights Into Cultivar Evaluation, Conservation, and Utilization
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ecosystem Services Changes Between 2000 and 2015 in the Loess Plateau, China: a Response to Ecological Restoration
RESEARCH ARTICLE Ecosystem services changes between 2000 and 2015 in the Loess Plateau, China: A response to ecological restoration Dan Wu1, Changxin Zou1, Wei Cao2*, Tong Xiao3, Guoli Gong4 1 Nanjing Institute of Environmental Sciences, Ministry of Environmental Protection, Nanjing, China, 2 Key Laboratory of Land Surface Pattern and Simulation, Institute of Geographic Sciences and Natural Resources Research, CAS, Beijing, China, 3 Satellite Environment Center, Ministry of Environmental Protection, Beijing, China, 4 Shanxi Academy of Environmental Planning, Taiyuan, China a1111111111 a1111111111 * [email protected] a1111111111 a1111111111 a1111111111 Abstract The Loess Plateau of China is one of the most severe soil and water loss areas in the world. Since 1999, the Grain to Green Program (GTGP) has been implemented in the region. This OPEN ACCESS study aimed to analyze spatial and temporal variations of ecosystem services from 2000 to Citation: Wu D, Zou C, Cao W, Xiao T, Gong G 2015 to assess the effects of the GTGP, including carbon sequestration, water regulation, (2019) Ecosystem services changes between 2000 soil conservation and sand fixation. During the study period, the area of forest land and and 2015 in the Loess Plateau, China: A response grassland significantly expanded, while the area of farmland decreased sharply. Ecosystem to ecological restoration. PLoS ONE 14(1): services showed an overall improvement with localized deterioration. Carbon sequestration, e0209483. https://doi.org/10.1371/journal. pone.0209483 water regulation and soil conservation increased substantially. Sand fixation showed a decreasing trend mainly because of decreased wind speeds. There were synergies Editor: Debjani Sihi, Oak Ridge National Laboratory, UNITED STATES between carbon sequestration and water regulation, and tradeoffs between soil conserva- tion and sand fixation. -

(Leech, 1890) (Lepidoptera: Hesperiidae) with Description of Female Genitalia and Taxonomic Notes
© Entomologica Fennica. 31 August 2016 Distribution of Onryza maga (Leech, 1890) (Lepidoptera: Hesperiidae) with description of female genitalia and taxonomic notes Guoxi Xue, Yufei Li, Zihao Liu, Meng Li & Yingdang Ren Xue, G. X., Li, Y.F., Liu, Z. H., Li, M. & Ren, Y.D. 2016: Distribution of Onryza maga (Leech, 1890) (Lepidoptera: Hesperiidae) with description of female geni- talia and taxonomic notes. — Entomol. Fennica 27: 70–76. For more than twenty years, Hainan, Vietnam, Myanmar, Thailand, Malaysia, Singapore and Indonesia have been erroneously reported in Chinese literature as belonging to the distribution range of Onryza maga (Leech 1890). Based upon a careful survey of specimens and relevant literature, these regions are omitted from the known range of this species. Onryza maga maga is found from northeast Guizhou, south Henan and Qinling-Daba Mountains in Shaanxi of China, its oc- currence in Hunan is confirmed. The adults are redescribed and the variability of wing patterns is discussed. Female genitalia are illustrated and described for the first time. Some biological information and an updated distribution map of the species are provided. G. X. Xue & M. Li, School of Food and Bioengineering, Zhengzhou University of Light Industry, No. 5 Dongfeng Road, Zhengzhou, Henan, 450002, P. R. China; Corresponding author’s e-mail: [email protected] Y. F. Li, School of Medicine, Xi’an Jiaotong University, No. 76 Yanta West Road, Xi’an, Shaanxi, 710061, P. R. China Z. H. Liu, School of Physics, University of Science and Technology of China, No. 96 Jinzhai Road, Hefei, Anhui, 230026, P. R. China Y. D. -

Preparing the Shaanxi-Qinling Mountains Integrated Ecosystem Management Project (Cofinanced by the Global Environment Facility)
Technical Assistance Consultant’s Report Project Number: 39321 June 2008 PRC: Preparing the Shaanxi-Qinling Mountains Integrated Ecosystem Management Project (Cofinanced by the Global Environment Facility) Prepared by: ANZDEC Limited Australia For Shaanxi Province Development and Reform Commission This consultant’s report does not necessarily reflect the views of ADB or the Government concerned, and ADB and the Government cannot be held liable for its contents. (For project preparatory technical assistance: All the views expressed herein may not be incorporated into the proposed project’s design. FINAL REPORT SHAANXI QINLING BIODIVERSITY CONSERVATION AND DEMONSTRATION PROJECT PREPARED FOR Shaanxi Provincial Government And the Asian Development Bank ANZDEC LIMITED September 2007 CURRENCY EQUIVALENTS (as at 1 June 2007) Currency Unit – Chinese Yuan {CNY}1.00 = US $0.1308 $1.00 = CNY 7.64 ABBREVIATIONS ADB – Asian Development Bank BAP – Biodiversity Action Plan (of the PRC Government) CAS – Chinese Academy of Sciences CASS – Chinese Academy of Social Sciences CBD – Convention on Biological Diversity CBRC – China Bank Regulatory Commission CDA - Conservation Demonstration Area CNY – Chinese Yuan CO – company CPF – country programming framework CTF – Conservation Trust Fund EA – Executing Agency EFCAs – Ecosystem Function Conservation Areas EIRR – economic internal rate of return EPB – Environmental Protection Bureau EU – European Union FIRR – financial internal rate of return FDI – Foreign Direct Investment FYP – Five-Year Plan FS – Feasibility -

GIS Assessment of the Status of Protected Areas in East Asia
CIS Assessment of the Status of Protected Areas in East Asia Compiled and edited by J. MacKinnon, Xie Yan, 1. Lysenko, S. Chape, I. May and C. Brown March 2005 IUCN V 9> m The World Conservation Union UNEP WCMC Digitized by the Internet Archive in 20/10 with funding from UNEP-WCMC, Cambridge http://www.archive.org/details/gisassessmentofs05mack GIS Assessment of the Status of Protected Areas in East Asia Compiled and edited by J. MacKinnon, Xie Yan, I. Lysenko, S. Chape, I. May and C. Brown March 2005 UNEP-WCMC IUCN - The World Conservation Union The designation of geographical entities in this book, and the presentation of the material, do not imply the expression of any opinion whatsoever on the part of UNEP, UNEP-WCMC, and IUCN concerning the legal status of any country, territory, or area, or of its authorities, or concerning the delimitation of its frontiers or boundaries. UNEP-WCMC or its collaborators have obtained base data from documented sources believed to be reliable and made all reasonable efforts to ensure the accuracy of the data. UNEP-WCMC does not warrant the accuracy or reliability of the base data and excludes all conditions, warranties, undertakings and terms express or implied whether by statute, common law, trade usage, course of dealings or otherwise (including the fitness of the data for its intended use) to the fullest extent permitted by law. The views expressed in this publication do not necessarily reflect those of UNEP, UNEP-WCMC, and IUCN. Produced by: UNEP World Conservation Monitoring Centre and IUCN, Gland, Switzerland and Cambridge, UK Cffti IUCN UNEP WCMC The World Conservation Union Copyright: © 2005 UNEP World Conservation Monitoring Centre Reproduction of this publication for educational or other non-commercial purposes is authorized without prior written permission from the copyright holder provided the source is fully acknowledged. -

Genomic Surveillance: Inside China's DNA Dragnet
Genomic surveillance Inside China’s DNA dragnet Emile Dirks and James Leibold Policy Brief Report No. 34/2020 About the authors Emile Dirks is a PhD candidate in political science at the University of Toronto. Dr James Leibold is an Associate Professor and Head of the Department of Politics, Media and Philosophy at La Trobe University and a non-resident Senior Fellow at ASPI. Acknowledgements The authors would like to thank Danielle Cave, Derek Congram, Victor Falkenheim, Fergus Hanson, William Goodwin, Bob McArthur, Yves Moreau, Kelsey Munro, Michael Shoebridge, Maya Wang and Sui-Lee Wee for valuable comments and suggestions with previous drafts of this report, and the ASPI team (including Tilla Hoja, Nathan Ruser and Lin Li) for research and production assistance with the report. ASPI is grateful to the Institute of War and Peace Reporting and the US State Department for supporting this research project. What is ASPI? The Australian Strategic Policy Institute was formed in 2001 as an independent, non-partisan think tank. Its core aim is to provide the Australian Government with fresh ideas on Australia’s defence, security and strategic policy choices. ASPI is responsible for informing the public on a range of strategic issues, generating new thinking for government and harnessing strategic thinking internationally. ASPI International Cyber Policy Centre ASPI’s International Cyber Policy Centre (ICPC) is a leading voice in global debates on cyber and emerging technologies and their impact on broader strategic policy. The ICPC informs public debate and supports sound public policy by producing original empirical research, bringing together researchers with diverse expertise, often working together in teams. -

Hubei Shennongjia
ASIA / PACIFIC HUBEI SHENNONGJIA CHINA Laojunshan Component of the property - © IUCN Bruce Jefferies China - Hubei Shennongjia WORLD HERITAGE NOMINATION – IUCN TECHNICAL EVALUATION HUBEI SHENNONGJIA (CHINA) – ID 1509 IUCN RECOMMENDATION TO WORLD HERITAGE COMMITTEE: To inscribe the property under natural criteria. Key paragraphs of Operational Guidelines: Paragraph 77: Nominated property meets World Heritage criteria. Paragraph 78: Nominated property meets integrity and protection and management requirements. 1. DOCUMENTATION S. and Hong Qian. Global Significance of Plant Diversity in China. In The Plants of China: A a) Date nomination received by IUCN: 16 March Companion to the Flora of China (2015). Huang, J. H., 2015 Chen, J.H., Ying, J.S., and Ke‐Ping M. Features and distribution patterns of Chinese endemic seed plant b) Additional information officially requested from species. Journal of Systematics and Evolution 49, no. and provided by the State Party: On 6 September 2 (2011): 81-94. Li, Y. (2004). The effect of forest 2015, the State Party responded to issues which arose clear-cutting on habitat use in Sichuan snub-nosed during the course of the IUCN field evaluation mission. monkey (Rhinopithecus roxellana) in Shennongjia The letter, with accompanying maps, addressed a Nature Reserve, China. Primates 45.1 69-72.. López- range of issues and confirmed extensions to the Pujol, J., et al. (2011). Mountains of Southern China as nominated area and buffer zone in the Badong County “plant museums” and “plant cradles”: evolutionary and area. Following the IUCN World Heritage Panel a conservation insights. Mountain Research and progress report was sent to the State Party on 16 Development,31(3), 261-269. -
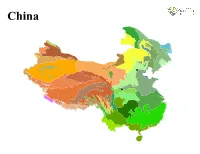
R Graphics Output
China China LEGEND Previously sampled Malaise trap site Ecoregion Alashan Plateau semi−desert North Tibetan Plateau−Kunlun Mountains alpine desert Altai alpine meadow and tundra Northeast China Plain deciduous forests Altai montane forest and forest steppe Northeast Himalayan subalpine conifer forests Altai steppe and semi−desert Northern Indochina subtropical forests Amur meadow steppe Northern Triangle subtropical forests Bohai Sea saline meadow Northwestern Himalayan alpine shrub and meadows Central China Loess Plateau mixed forests Nujiang Langcang Gorge alpine conifer and mixed forests Central Tibetan Plateau alpine steppe Ordos Plateau steppe Changbai Mountains mixed forests Pamir alpine desert and tundra Changjiang Plain evergreen forests Qaidam Basin semi−desert Da Hinggan−Dzhagdy Mountains conifer forests Qilian Mountains conifer forests Daba Mountains evergreen forests Qilian Mountains subalpine meadows Daurian forest steppe Qin Ling Mountains deciduous forests East Siberian taiga Qionglai−Minshan conifer forests Eastern Gobi desert steppe Rock and Ice Eastern Himalayan alpine shrub and meadows Sichuan Basin evergreen broadleaf forests Eastern Himalayan broadleaf forests South China−Vietnam subtropical evergreen forests Eastern Himalayan subalpine conifer forests Southeast Tibet shrublands and meadows Emin Valley steppe Southern Annamites montane rain forests Guizhou Plateau broadleaf and mixed forests Suiphun−Khanka meadows and forest meadows Hainan Island monsoon rain forests Taklimakan desert Helanshan montane conifer forests -

Pn Wave Velocity and Anisotropy Underneath the Central
Journal of Asian Earth Sciences 184 (2019) 103941 Contents lists available at ScienceDirect Journal of Asian Earth Sciences journal homepage: www.elsevier.com/locate/jseaes Pn wave velocity and anisotropy underneath the central segment of the North-South Seismic Belt in China T ⁎ Guangbao Dua,b, Qingju Wua, Xuemei Zhangb, , Jing Hec, Liye Zoub, Yangyang Fengd, Jie Liub, Fabio Romanellie a Key Laboratory of Seismic Observation and Geophysical Imaging, Institute of Geophysics, China Earthquake Administration, Beijing 100081, China b China Earthquake Networks Center, Beijing 100045, China c Key Laboratory of Crustal Dynamics, Institute of Crustal Dynamics, China Earthquake Administration, Beijing 100085, China d The Inner Mongolia Autonomous Region Seismological Bureau, Hohhot 010051, China e Department of Mathematics and Geosciences, University of Trieste, Trieste 34127 Italy ARTICLE INFO ABSTRACT Keywords: We present a Pn wave velocity and anisotropy model of the central segment of the North-South Seismic Belt in Pn wave China, where there are numerous stable basins and active faults, making this segment attractive for extensive Velocity heterogeneity studies. The model was obtained by a tomographic analysis of 49,973 Pn wave phase readings collected by the Anisotropy China Earthquake Networks Center and temporary stations in Yunnan and Sichuan. The tomographic velocity Uppermost mantle model shows that the average Pn wave velocity is 8.06 km/s; prominent high-velocity (high-V) anomalies are Central segment of the North-South Seismic visible under the Sichuan Basin, the Zoige Basin and the Ordos block, which clearly outline their tectonic Belt margins. A pronounced low-velocity (low-V) zone is observed from the Songpan-Ganzi block to the Chuan-Dian and Daliangshan blocks, suggesting the presence of hot material upwelling. -

Analysis of the Spatial-Temporal Change of the Vegetation Index in the Upper Reach of Han River Basin in 2000–2016
Innovative water resources management – understanding and balancing interactions between humankind and nature Proc. IAHS, 379, 287–292, 2018 https://doi.org/10.5194/piahs-379-287-2018 Open Access © Author(s) 2018. This work is distributed under the Creative Commons Attribution 4.0 License. Analysis of the spatial-temporal change of the vegetation index in the upper reach of Han River Basin in 2000–2016 Jinkai Luan1, Dengfeng Liu1,2, Lianpeng Zhang1, Qiang Huang1, Jiuliang Feng3, Mu Lin4, and Guobao Li5 1State Key Laboratory of Eco-hydraulics in Northwest Arid Region of China, School of Water Resources and Hydropower, Xi’an University of Technology, Xi’an 710048, China 2Department of Land Resources and Environmental Sciences, Montana State University, Bozeman, MT 59717, USA 3Shanxi Provincal Water and Soil Conservation and Ecological Environment Construction Center, Taiyuan 030002, China 4School of statistics and Mathematics, Central University of Finance and Economics, Beijing 100081, China 5Work team of hydraulic of Yulin City, Yulin 719000, China Correspondence: Dengfeng Liu ([email protected]) Received: 29 December 2017 – Revised: 25 March 2018 – Accepted: 26 March 2018 – Published: 5 June 2018 Abstract. Han River is the water source region of the middle route of South-to-North Water Diversion in China and the ecological projects were implemented since many years ago. In order to monitor the change of vegetation in Han River and evaluate the effect of ecological projects, it is needed to reveal the spatial-temporal change of the vegetation in the upper reach of Han River quantitatively. The study is based on MODIS/Terra NDVI remote sensing data, and analyzes the spatial-temporal changes of the NDVI in August from 2000 to 2016 at pixel scale in the upper reach of Han River Basin. -

Dear Editor and Reviewers
1. Page 3652, lines 10-16: The table containing the site information is well-done, but within the manuscript it would be good to include the elevation range of the sampling sites. >> Revised as suggested - the elevation of all sampling sites was added to the table. The elevation ranges from 169 m to 661m above sea level. 2. P. 3652, l. 19: Is there any idea of the inter-annual variation in rainfall or temperature in this region? Perhaps error of some type here. Also, are there any present temperature/rainfall trends seen during this time period? >> Yes, we have added the inter-annual variation and presented rainfall and air temperature in this region based on seven meteorological stations located in the respective counties in this region (Shiyan City, Danjiangkou City, Yun County, Yunxi County, Fang County, Zhuxi County and Zhushan County) from 1961 to 2009. This information is given in the publication of Zhu et al., 2010. The data show that there is little interannual variation in rainfall and temperature for these sites (coefficient of variations of 5% and 1%). Present temperature / rainfall trends were (not) observed in the experiment year. All this information was added to the M+M section of the revised version. 3. P. 3652, l. 22: Where did the measure of sunshine hours come from? >> It comes from the reference of Zhu et al., 2010. We added this citation to the reference list. 4. P. 3653, l. 4-8: The description of the site selection process is lacking. How did “experienced staff members” select this sites? Where the selections random? Soil type and elevation have the potential to greatly influence the outcomes of these findings, the manner in which these site characteristics were consider in selecting study sites is crucial and thus this area of the manuscript needs further explication. -

A New Species of Odorrana (Anura, Ranidae) from Hunan Province, China
ZooKeys 1024: 91–115 (2021) A peer-reviewed open-access journal doi: 10.3897/zookeys.1024.56399 RESEarch arTICLE https://zookeys.pensoft.net Launched to accelerate biodiversity research A new species of Odorrana (Anura, Ranidae) from Hunan Province, China Bing Zhang1, Yuan Li1, Ke Hu1, Pipeng Li2, Zhirong Gu3, Nengwen Xiao4, Daode Yang1 1 Institute of Wildlife Conservation, Central South University of Forestry and Technology, Changsha 410004, China 2 Institute of Herpetology, Shenyang Normal University, Shenyang 110034, China 3 Bureau of Hunan Badagongshan National Nature Reserve, Sangzhi 427100, China 4 State Environmental Protection Key Labo- ratory of Regional Eco-process and Function Assessment, Chinese Research Academy of Environmental Sciences, Beijing 100012, China Corresponding author: Daode Yang ([email protected]) Academic editor: A. Crottini | Received 12 July 2020 | Accepted 30 December 2020 | Published 15 March 2021 http://zoobank.org/756CA7F5-A4C1-4759-AB64-8C147F6C9A6A Citation: Zhang B, Li Y, Hu K, Li P, Gu Z, Xiao N, Yang D (2021) A new species of Odorrana (Anura, Ranidae) from Hunan Province, China. ZooKeys 1024: 91–115. https://doi.org/10.3897/zookeys.1024.56399 Abstract A new species, Odorrana sangzhiensis sp. nov., is described, based on five specimens from Sangzhi County, Zhangjiajie City, Hunan Province, China. Molecular phylogenetic analyses, based on mitochondrial 12S rRNA and 16S rRNA gene sequences, strongly support the new species as a monophyletic group nested into the O. schmackeri species complex. The new -

1. Brief Introduction to the Institute of Geology
-1- The Institute of Geology, Chinese Academy of Geological Sciences (CAGS) Preface The Institute of Geology, Chinese Academy of Geological Sciences (CAGS), is a national public scientific research institution and is mainly engaged in national fundamental, public, strategic and frontier geological survey and geoscientific research. Entering the new century, and in particular during the past 5 years, the Institute has made notable progress in scientific research, personnel training and international cooperation, with increasing cooperation and exchange activities, expanded fields of cooperation, abundant output of new research results, and an increased number of papers published in “Nature”, “Science” and other high-impact international scientific journals. In the light of this new situation and in order to publicize, in a timely manner, annual progress and achievements of the Institute to enhance its international reputation, an English version of the Institute’s Annual Report has been published since 2010. Similar to previous reports, the Annual Report 2015 includes the following 7 parts: (1) Introduction to the Institute of Geology, CAGS; (2) Ongoing Research Projects; (3) Research Achievements and Important Progress; (4) International Cooperation and Academic Exchange; (5) Important Academic Activities in 2015; (6) Postgraduate Education; (7) Publications. In order to avoid confusion in the meaning of Chinese and foreign names, all family names in this Report are capitalized. We express our sincere gratitude to colleagues of related research departments and centers of the Institute for their support and efforts in compiling this Report and providing related material – a written record of the hard work of the Institute’s scientific research personnel for the year 2015.