Regulation of Pre-Mrna Alternative Splicing by the RNA Processing
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification of the RNA Recognition Element of the RBPMS Family of RNA-Binding Proteins and Their Transcriptome-Wide Mrna Targets
Downloaded from rnajournal.cshlp.org on October 1, 2021 - Published by Cold Spring Harbor Laboratory Press Identification of the RNA recognition element of the RBPMS family of RNA-binding proteins and their transcriptome-wide mRNA targets THALIA A. FARAZI,1,5 CARL S. LEONHARDT,1,5 NEELANJAN MUKHERJEE,2 ALEKSANDRA MIHAILOVIC,1 SONG LI,3 KLAAS E.A. MAX,1 CINDY MEYER,1 MASASHI YAMAJI,1 PAVOL CEKAN,1 NICHOLAS C. JACOBS,2 STEFANIE GERSTBERGER,1 CLAUDIA BOGNANNI,1 ERIK LARSSON,4 UWE OHLER,2 and THOMAS TUSCHL1,6 1Laboratory of RNA Molecular Biology, Howard Hughes Medical Institute, The Rockefeller University, New York, New York 10065, USA 2Berlin Institute for Medical Systems Biology, Max Delbrück Center for Molecular Medicine, 13125 Berlin, Germany 3Biology Department, Duke University, Durham, North Carolina 27708, USA 4Institute of Biomedicine, The Sahlgrenska Academy, University of Gothenburg, Gothenburg, SE-405 30, Sweden ABSTRACT Recent studies implicated the RNA-binding protein with multiple splicing (RBPMS) family of proteins in oocyte, retinal ganglion cell, heart, and gastrointestinal smooth muscle development. These RNA-binding proteins contain a single RNA recognition motif (RRM), and their targets and molecular function have not yet been identified. We defined transcriptome-wide RNA targets using photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) in HEK293 cells, revealing exonic mature and intronic pre-mRNA binding sites, in agreement with the nuclear and cytoplasmic localization of the proteins. Computational and biochemical approaches defined the RNA recognition element (RRE) as a tandem CAC trinucleotide motif separated by a variable spacer region. Similar to other mRNA-binding proteins, RBPMS family of proteins relocalized to cytoplasmic stress granules under oxidative stress conditions suggestive of a support function for mRNA localization in large and/or multinucleated cells where it is preferentially expressed. -

Figure S1. Androgen Signaling Upregulates an Intronic
Figure S1. Androgen signaling upregulates an intronic polyadenylated EWSR1 isoform A) Gene expression correlation of EWSR1 and AR in 550 prostate cancer patients from the PRAD data set. B) EWSR1 polyadenylation site (PAS) information from PolyA_db v3.2. There are seven PAS for this gene and they are numbered starting from the 5’ end of the gene. C) Diagram of PAS locations at EWSR1 numbered according to table in A. D) Normalized read count for PAS #2, the PAS for ntEWS, from 3’ sequencing data across various cell types. E) RNA levels of PSA in VCaP, LNCaP, and LNCaP-AR cells treated with DMSO or 10nM R1881 for 24 hours. Expression is normalized to 18S and relative to the DMSO condition. The mean ± SEM for three replicates is shown. F) Immuno blot of ntEWS in PC3 cells overexpressing HA-ntEWS. G) Immunoblot of ntEWS in VCaP cells overexpressing vector alone or shRNAs targeting the 3’ UTR of ntEWS. Tubulin is used as a loading control for immunoblots. Figure S2. AR binding to Intron 5 of EWSR1 directly regulates ntEWS expression A) Gene tracks for AR binding in patient tumor and matched adjacent normal tissue at known AR enhancers. Order of tracks is consistent with Figure 2a. Figure S3. ntEWS promotes phenotypes related to oncogenesis A) Immunoblot of 3xHA tagged EWS isoforms expressed in PC3 cells. Tubulin is used as a loading control. B) MTT proliferation assay of PC3 isoform-expressing lines. Figure S4. The ntEWS alternative last exon encodes an alpha helical domain important for function A) IUPRED prediction of disorder of ntEWS (bottom) and EWS(1-355aa) (top). -

Large-Scale Analysis of Genome and Transcriptome Alterations in Multiple Tumors Unveils Novel Cancer-Relevant Splicing Networks
Downloaded from genome.cshlp.org on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks Endre Sebestyén1,*, Babita Singh1,*, Belén Miñana1,2, Amadís Pagès1, Francesca Mateo3, Miguel Angel Pujana3, Juan Valcárcel1,2,4, Eduardo Eyras1,4,5 1Universitat Pompeu Fabra, Dr. Aiguader 88, E08003 Barcelona, Spain 2Centre for Genomic Regulation, Dr. Aiguader 88, E08003 Barcelona, Spain 3Program Against Cancer Therapeutic Resistance (ProCURE), Catalan Institute of Oncology (ICO), Bellvitge Institute for Biomedical Research (IDIBELL), E08908 L’Hospitalet del Llobregat, Spain. 4Catalan Institution for Research and Advanced Studies, Passeig Lluís Companys 23, E08010 Barcelona, Spain *Equal contribution 5Correspondence to: [email protected] Keywords: alternative splicing, RNA binding proteins, splicing networks, cancer 1 Downloaded from genome.cshlp.org on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press Abstract Alternative splicing is regulated by multiple RNA-binding proteins and influences the expression of most eukaryotic genes. However, the role of this process in human disease, and particularly in cancer, is only starting to be unveiled. We systematically analyzed mutation, copy number and gene expression patterns of 1348 RNA-binding protein (RBP) genes in 11 solid tumor types, together with alternative splicing changes in these tumors and the enrichment of binding motifs in the alternatively spliced sequences. Our comprehensive study reveals widespread alterations in the expression of RBP genes, as well as novel mutations and copy number variations in association with multiple alternative splicing changes in cancer drivers and oncogenic pathways. Remarkably, the altered splicing patterns in several tumor types recapitulate those of undifferentiated cells. -

Roles of Splicing Factors in Hormone-Related Cancer Progression
International Journal of Molecular Sciences Review Roles of Splicing Factors in Hormone-Related Cancer Progression Toshihiko Takeiwa 1, Yuichi Mitobe 1, Kazuhiro Ikeda 1, Kuniko Horie-Inoue 1 and Satoshi Inoue 1,2,* 1 Division of Gene Regulation and Signal Transduction, Research Center for Genomic Medicine, Saitama Medical University, Hidaka, Saitama 350-1241, Japan; [email protected] (T.T.); [email protected] (Y.M.); [email protected] (K.I.); [email protected] (K.H.-I.) 2 Department of Systems Aging Science and Medicine, Tokyo Metropolitan Institute of Gerontology, Itabashi-ku, Tokyo 173-0015, Japan * Correspondence: [email protected]; Tel.: +81-3-3964-3241 Received: 8 February 2020; Accepted: 20 February 2020; Published: 25 February 2020 Abstract: Splicing of mRNA precursor (pre-mRNA) is a mechanism to generate multiple mRNA isoforms from a single pre-mRNA, and it plays an essential role in a variety of biological phenomena and diseases such as cancers. Previous studies have demonstrated that cancer-specific splicing events are involved in various aspects of cancers such as proliferation, migration and response to hormones, suggesting that splicing-targeting therapy can be promising as a new strategy for cancer treatment. In this review, we focus on the splicing regulation by RNA-binding proteins including Drosophila behavior/human splicing (DBHS) family proteins, serine/arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs) in hormone-related cancers, such as breast and prostate cancers. Keywords: DBHS family proteins; SR proteins; hnRNPs; breast cancer; prostate cancer 1. Introduction Splicing of mRNA precursors (pre-mRNAs) is an essential mechanism in the posttranscriptional regulation of gene expression. -

Regulation of Alternative Splicing by the Core Spliceosomal Machinery
Downloaded from genesdev.cshlp.org on September 23, 2021 - Published by Cold Spring Harbor Laboratory Press Regulation of alternative splicing by the core spliceosomal machinery Arneet L. Saltzman,1,2 Qun Pan,1 and Benjamin J. Blencowe1,2,3 1Banting and Best Department of Medical Research, The Donnelly Centre for Cellular and Biomolecular Research, University of Toronto, Toronto, Ontario M5S 3E1, Canada; 2Department of Molecular Genetics, University of Toronto, Toronto, Ontario M5S 1A8, Canada Alternative splicing (AS) plays a major role in the generation of proteomic diversity and in gene regulation. However, the role of the basal splicing machinery in regulating AS remains poorly understood. Here we show that the core snRNP (small nuclear ribonucleoprotein) protein SmB/B9 self-regulates its expression by promoting the inclusion of a highly conserved alternative exon in its own pre-mRNA that targets the spliced transcript for nonsense-mediated mRNA decay (NMD). Depletion of SmB/B9 in human cells results in reduced levels of snRNPs and a striking reduction in the inclusion levels of hundreds of additional alternative exons, with comparatively few effects on constitutive exon splicing levels. The affected alternative exons are enriched in genes encoding RNA processing and other RNA-binding factors, and a subset of these exons also regulate gene expression by activating NMD. Our results thus demonstrate a role for the core spliceosomal machinery in controlling an exon network that appears to modulate the levels of many RNA processing factors. [Keywords: alternative splicing; Sm proteins; snRNP; autoregulation; NMD; exon network] Supplemental material is available for this article. Received October 20, 2010; revised version accepted January 4, 2011. -

A Heroin Addiction Severity-Associated Intronic Single
11048 • The Journal of Neuroscience, August 13, 2014 • 34(33):11048–11066 Cellular/Molecular A Heroin Addiction Severity-Associated Intronic Single Nucleotide Polymorphism Modulates Alternative Pre-mRNA Splicing of the Opioid Receptor Gene OPRM1 via hnRNPH Interactions Jin Xu,1* Zhigang Lu,1* Mingming Xu,1 Ling Pan,1 Yi Deng,1 Xiaohu Xie,3 Huifen Liu,3 Shixiong Ding,4 Yasmin L. Hurd,5 X Gavril W. Pasternak,1 X Robert J. Klein,2 Luca Cartegni,6 X Wenhua Zhou,3† and Ying-Xian Pan1† 1Department of Neurology and the Molecular Pharmacology & Chemistry Program, and 2Department of Medicine, Memorial Sloan Kettering Cancer Center, New York, New York 10065, 3Laboratory of Behavioral Neuroscience, Ningbo Addiction Research and Treatment Center, School of Medicine, Ningbo University, Ningbo, Zhejiang 315211, People’s Republic of China, 4Laboratory of Clinical Biochemistry, Ningbo No. 2 Hospital, Zhejiang 315100, People’s Republic of China, 5Departments of Psychiatry and Neuroscience, Icahn School of Medicine at Mount Sinai, New York, New York 10029, and 6Susan Lehman Cullman Laboratory for Cancer Research, Ernest Mario School of Pharmacy, Rutgers, The State University of New Jersey, Piscataway, New Jersey 08854 Single nucleotide polymorphisms (SNPs) in the OPRM1 gene have been associated with vulnerability to opioid dependence. The current study identifies an association of an intronic SNP (rs9479757) with the severity of heroin addiction among Han-Chinese male heroin addicts. Individual SNP analysis and haplotype-based analysis with additional SNPs in the OPRM1 locus showed that mild heroin addiction was associated with the AG genotype, whereas severe heroin addiction was associated with the GG genotype. -

Biology of the Mrna Splicing Machinery and Its Dysregulation in Cancer Providing Therapeutic Opportunities
International Journal of Molecular Sciences Review Biology of the mRNA Splicing Machinery and Its Dysregulation in Cancer Providing Therapeutic Opportunities Maxime Blijlevens †, Jing Li † and Victor W. van Beusechem * Medical Oncology, Amsterdam UMC, Cancer Center Amsterdam, Vrije Universiteit Amsterdam, de Boelelaan 1117, 1081 HV Amsterdam, The Netherlands; [email protected] (M.B.); [email protected] (J.L.) * Correspondence: [email protected]; Tel.: +31-2044-421-62 † Shared first author. Abstract: Dysregulation of messenger RNA (mRNA) processing—in particular mRNA splicing—is a hallmark of cancer. Compared to normal cells, cancer cells frequently present aberrant mRNA splicing, which promotes cancer progression and treatment resistance. This hallmark provides opportunities for developing new targeted cancer treatments. Splicing of precursor mRNA into mature mRNA is executed by a dynamic complex of proteins and small RNAs called the spliceosome. Spliceosomes are part of the supraspliceosome, a macromolecular structure where all co-transcriptional mRNA processing activities in the cell nucleus are coordinated. Here we review the biology of the mRNA splicing machinery in the context of other mRNA processing activities in the supraspliceosome and present current knowledge of its dysregulation in lung cancer. In addition, we review investigations to discover therapeutic targets in the spliceosome and give an overview of inhibitors and modulators of the mRNA splicing process identified so far. Together, this provides insight into the value of targeting the spliceosome as a possible new treatment for lung cancer. Citation: Blijlevens, M.; Li, J.; van Beusechem, V.W. Biology of the Keywords: alternative splicing; splicing dysregulation; splicing factors; NSCLC mRNA Splicing Machinery and Its Dysregulation in Cancer Providing Therapeutic Opportunities. -

Principles of RNA Processing from Analysis of Enhanced CLIP Maps for 150 RNA Binding Proteins Eric L
Van Nostrand et al. Genome Biology (2020) 21:90 https://doi.org/10.1186/s13059-020-01982-9 RESEARCH Open Access Principles of RNA processing from analysis of enhanced CLIP maps for 150 RNA binding proteins Eric L. Van Nostrand1,2, Gabriel A. Pratt1,2, Brian A. Yee1,2, Emily C. Wheeler1,2, Steven M. Blue1,2, Jasmine Mueller1,2, Samuel S. Park1,2, Keri E. Garcia1,2, Chelsea Gelboin-Burkhart1,2, Thai B. Nguyen1,2, Ines Rabano1,2, Rebecca Stanton1,2, Balaji Sundararaman1,2, Ruth Wang1,2, Xiang-Dong Fu1,2, Brenton R. Graveley3* and Gene W. Yeo1,2* Abstract Background: A critical step in uncovering rules of RNA processing is to study the in vivo regulatory networks of RNA binding proteins (RBPs). Crosslinking and immunoprecipitation (CLIP) methods enable mapping RBP targets transcriptome-wide, but methodological differences present challenges to large-scale analysis across datasets. The development of enhanced CLIP (eCLIP) enabled the mapping of targets for 150 RBPs in K562 and HepG2, creating a unique resource of RBP interactomes profiled with a standardized methodology in the same cell types. Results: Our analysis of 223 eCLIP datasets reveals a range of binding modalities, including highly resolved positioning around splicing signals and mRNA untranslated regions that associate with distinct RBP functions. Quantification of enrichment for repetitive and abundant multicopy elements reveals 70% of RBPs have enrichment for non-mRNA element classes, enables identification of novel ribosomal RNA processing factors and sites, and suggests that association with retrotransposable elements reflects multiple RBP mechanisms of action. Analysis of spliceosomal RBPs indicates that eCLIP resolves AQR association after intronic lariat formation, enabling identification of branch points with single-nucleotide resolution, and provides genome-wide validation for a branch point-based scanning model for 3′ splice site recognition. -

Sequence, Structure and Context Preferences of Human RNA
bioRxiv preprint doi: https://doi.org/10.1101/201996; this version posted October 12, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Sequence, Structure and Context Preferences of Human RNA Binding Proteins Daniel Dominguez§,1, Peter Freese§,2, Maria Alexis§,2, Amanda Su1, Myles Hochman1, Tsultrim Palden1, Cassandra Bazile1, Nicole J Lambert1, Eric L Van Nostrand3,4, Gabriel A. Pratt3,4,5, Gene W. Yeo3,4,6,7, Brenton R. Graveley8, Christopher B. Burge1,9,* 1. Department of Biology, MIT, Cambridge MA 2. Program in Computational and Systems Biology, MIT, Cambridge MA 3. Department of Cellular and Molecular Medicine, University of California at San Diego, La Jolla, CA 4. Institute for Genomic Medicine, University of California at San Diego, La Jolla, CA 5. Bioinformatics and Systems Biology Graduate Program, University of California San Diego, La Jolla, CA 6. Department of Physiology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore 7. Molecular Engineering Laboratory. A*STAR, Singapore 8. Department of Genetics and Genome Sciences, Institute for Systems Genomics, Univ. Connecticut Health, Farmington, CT 9. Department of Biological Engineering, MIT, Cambridge MA * Address correspondence to: [email protected] 1 of 61 bioRxiv preprint doi: https://doi.org/10.1101/201996; this version posted October 12, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Knockdown of Hnrnpa0, a Del(5Q) Gene, Alters Myeloid Cell Fate In
Myelodysplastic Syndromes SUPPLEMENTARY APPENDIX Knockdown of Hnrnpa0 , a del(5q) gene, alters myeloid cell fate in murine cells through regulation of AU-rich transcripts David J. Young, 1 Angela Stoddart, 2 Joy Nakitandwe, 3 Shann-Ching Chen, 3 Zhijian Qian, 4 James R. Downing, 3 and Michelle M. Le Beau 2 1Department of Pediatrics, Division of Oncology, Johns Hopkins University, Baltimora, MD; 2Department of Medicine and the Compre - hensive Cancer Center, University of Chicago, IL; 3St. Jude Children's Research Hospital, Memphis, Tennessee; and 4University of Illi - nois Cancer Center, Chicago, IL, USA DJY and AS equally contributed to this work. ©2014 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol.2013.098657 Manuscript received on September 25, 2013. Manuscript accepted on February 13, 2014. Correspondence: [email protected] Supplementary Materials for D. Young et al. Purification of hematopoietic populations from mice. Cells from the spleens, thymi, and bone marrow of C57BL/6J mice were harvested as appropriate for each population. For primitive populations including Lin–Sca-1+Kit+ (LSK), common lymphoid (CLP) and myeloid (CMP) progenitors, and granulocyte- monocyte progenitors (GMP), the cells were depleted of mature cells using the Mouse Hematopoietic Progenitor Cell Enrichment Kit (StemCell Technologies). The cells were stained for appropriate lineage markers, as described in Supplementary Figure S1, and sorted using a FACSAria fluorescence activated cell sorter (BD Biosciences). Real-time RT-PCR analysis Total RNA was purified from cells using Stat-60 (Tel-Test), according to the manufacturer’s protocols. First-strand cDNA was synthesized using SuperScript III SuperMix for qRT-PCR (Invitrogen) containing both random hexamers and oligo(dT)20 for priming. -

Global Analysis of Physical and Functional RNA Targets of Hnrnp L Reveals Distinct Sequence and Epigenetic Features of Repressed and Enhanced Exons
Global analysis of physical and functional RNA targets of hnRNP L reveals distinct sequence and epigenetic features of repressed and enhanced exons BRIAN S. COLE,1,2 IULIA TAPESCU,1,2 SAMUEL J. ALLON,1,2 MICHAEL J. MALLORY,1,2 JINSONG QIU,3 ROBERT J. LAKE,1,2,4 HUA-YING FAN,1,2,4 XIANG-DONG FU,3 and KRISTEN W. LYNCH1,2 1Department of Biochemistry and Biophysics, 2Department of Genetics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania 19104, USA 3Department of Cell and Molecular Medicine, University of California San Diego, San Diego, California 92093, USA 4Epigenetics Program, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania 19104, USA ABSTRACT HnRNP L is a ubiquitous splicing-regulatory protein that is critical for the development and function of mammalian T cells. Previous work has identified a few targets of hnRNP L-dependent alternative splicing in T cells and has described transcriptome-wide association of hnRNP L with RNA. However, a comprehensive analysis of the impact of hnRNP L on mRNA expression remains lacking. Here we use next-generation sequencing to identify transcriptome changes upon depletion of hnRNP L in a model T-cell line. We demonstrate that hnRNP L primarily regulates cassette-type alternative splicing, with minimal impact of hnRNP L depletion on transcript abundance, intron retention, or other modes of alternative splicing. Strikingly, we find that binding of hnRNP L within or flanking an exon largely correlates with exon repression by hnRNP L. In contrast, exons that are enhanced by hnRNP L generally lack proximal hnRNP L binding. -
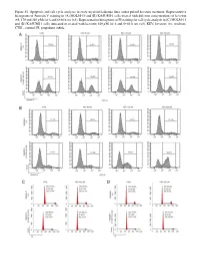
Figure S1. Apoptosis and Cell Cycle Analyses in Acute Myeloid Leukemia Lines Under Pulsed Kevetrin Treatment
Figure S1. Apoptosis and cell cycle analyses in acute myeloid leukemia lines under pulsed kevetrin treatment. Representative histograms of Annexin V staining in (A) MOLM‑13 and (B) KASUMI‑1 cells treated with different concentrations of kevetrin (85, 170 and 340 µM) for 6 and 6+66 h wo (x3). Representative histograms of PI staining for cell cycle analysis in (C) MOLM‑13 and (D) KASUMI‑1 cells untreated or treated with kevetrin 340 µM for 6 and 6+66 h wo (x3). KEV, kevetrin; wo, washout; CTRL, control; PI, propidium iodide. Figure S2. Viability of acute myeloid leukemia cell lines after 72 h of kevetrin treatment. Viability of OCI‑AML3, MOLM‑13, KASUMI‑1 and NOMO‑1 cell lines treated with different concentrations of kevetrin (85, 170 and 340 µM) for 72 h. Values represent the mean ± standard deviation of three biological replicates. **P<0.01, ***P<0.001. Figure S3. Apoptosis analysis in acute myeloid leukemia cell lines under 24 h of continuous kevetrin treatment. Representative histograms of Annexin V staining in OCI‑AML3, MOLM‑13, KASUMI‑1 and NOMO‑1 cells treated with different concentra‑ tions of kevetrin (85, 170 and 340 µM) for 24 h. KEV, kevetrin; CTRL, control. Figure S4. Apoptosis analysis in acute myeloid leukemia cell lines under 48 h of continuous kevetrin treatment. Representative histograms of Annexin V staining in OCI‑AML3, MOLM‑13, KASUMI‑1 and NOMO‑1 cells treated with different concentra‑ tions of kevetrin (85, 170 and 340 µM) for 48 h. KEV, kevetrin; CTRL, control. Figure S5. Apoptosis analyses in acute myeloid leukemia cell lines under continuous kevetrin treatment.