Identification of Molecular Markers Linked to the Trait of Natural Astringency Loss of Japanese Persimmon (Diospyros Kaki) Fruit
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Differences in the Phenolic Profile by UPLC Coupled to High Resolution
antioxidants Article Differences in the Phenolic Profile by UPLC Coupled to High Resolution Mass Spectrometry and Antioxidant Capacity of Two Diospyros kaki Varieties Adelaida Esteban-Muñoz 1,2,*, Silvia Sánchez-Hernández 1,2, Cristina Samaniego-Sánchez 1,3 , Rafael Giménez-Martínez 1,3 and Manuel Olalla-Herrera 1,3 1 Departamento de Nutrición y Bromatología, Universidad de Granada, 18071 Granada, Spain; [email protected] (S.S.-H.); [email protected] (C.S.-S.); [email protected] (R.G.-M.); [email protected] (M.O.-H.) 2 Programme in Nutrition and Food Science, University of Granada, 18071 Granada, Spain 3 Biosanitary Research Institute, IBS, 18071 Granada, Spain * Correspondence: [email protected]; Tel.: +34-958-243-863 Abstract: Background: phenolic compounds are bioactive chemical species derived from fruits and vegetables, with a plethora of healthy properties. In recent years, there has been a growing inter- est in persimmon (Diospyros kaki L.f.) due to the presence of many different classes of phenolic compounds. However, the analysis of individual phenolic compounds is difficult due to matrix interferences. Methods: the aim of this research was the evaluation of individual phenolic compounds and antioxidant capacity of the pulp of two varieties of persimmon (Rojo Brillante and Triumph) by an improved extraction procedure together with a UPLC-Q-TOF-MS platform. Results: the phenolic compounds composition of persimmon was characterized by the presence of hydroxybenzoic and hy- droxycinnamic acids, hydroxybenzaldehydes, dihydrochalcones, tyrosols, flavanols, flavanones, and Citation: Esteban-Muñoz, A.; flavonols. A total of 31 compounds were identified and 17 compounds were quantified. Gallic acid was Sánchez-Hernández, S.; Rojo Brillante Samaniego-Sánchez, C.; the predominant phenolic compounds found in the variety (0.953 mg/100 g) whereas the Giménez-Martínez, R.; concentration of p-hydroxybenzoic acid was higher in the Triumph option (0.119 mg/100 g). -
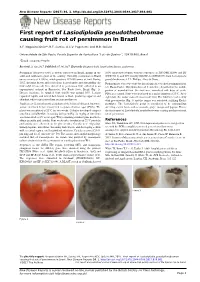
First Report of Lasiodiplodia Pseudotheobromae Causing Fruit Rot of Persimmon in Brazil
New Disease Reports (2017) 36, 1. http://dx.doi.org/10.5197/j.2044-0588.2017.036.001 First report of Lasiodiplodia pseudotheobromae causing fruit rot of persimmon in Brazil A.F. Nogueira Júnior*, R.F. Santos, A.C.V. Pagenotto and M.B. Spósito Universidade de São Paulo, Escola Superior de Agricultura “Luiz de Queiroz”, 13418-900, Brazil *E-mail: [email protected] Received: 23 Jun 2017. Published: 07 Jul 2017. Keywords: Diospyros kaki, fungal plant disease, postharvest Persimmon (Diospyros kaki) is widely cultivated in Brazil, mainly in the 100% nucleotide identity with the sequences of ITS (NR111264) and BT south and southeast regions of the country. Currently, persimmon in Brazil (EU673111), and 99% identity with EF-1α (EF622057) from Lasiodiplodia covers an area of 8,300 ha, which produces 182,000 tonnes of fruit. During pseudotheobromae A.J.L. Phillips, Alves & Crous. 2015, irregular brown and soft lesions located under and surrounding the Pathogenicity tests were done by inoculating six detached persimmon fruit fruit calyx (stem-end) were observed in persimmon fruit collected in an (cv. Rama Forte). Mycelium discs of 8 mm were deposited in the middle experimental orchard in Piracicaba, Sao Paulo State, Brazil (Fig. 1). portion of wounded fruit. Six fruit were inoculated with discs of sterile Disease incidence in sampled fruit (n=50) was around 10%. Lesions PDA as a control. Fruit were incubated in a moist chamber at 25°C. After expanded rapidly and turned dark brown to black producing apparent and eight days, the isolate caused lesions in all fruit. The fruit were covered by abundant white to grey mycelium on fruit postharvest. -

Chemical Composition of the Essential Oil of Diospyros Wallichii King & Gamble (Ebenaceae) Wan Mohd Nuzul Hakimi Wan Salleh1, * and Shamsul Khamis2
Nat. Volatiles & Essent. Oils, 2020; 7(3): 12-17 Salleh & Khamis DOI: 10.37929/nveo.746965 RESEARCH ARTICLE Chemical composition of the essential oil of Diospyros wallichii King & Gamble (Ebenaceae) Wan Mohd Nuzul Hakimi Wan Salleh1, * and Shamsul Khamis2 1Department of Chemistry, Faculty of Science and Mathematics, University Pendidikan Sultan Idris (UPSI), 35900 Tanjung Malim, Perak, MALAYSIA 2School of Environmental and Natural Sciences, Faculty of Science and Technology, Universiti Kebangsaan Malaysia, 43600 Bangi, Selangor, MALAYSIA *Corresponding author. Email: [email protected] Submitted: 02.06.2020; Accepted: 18.08.2020 Abstract The chemical composition of the essential oil from the leaves of Diospyros wallichii (Ebenaceae) growing in Malaysia was investigated for the first time. The essential oil was obtained by hydrodistillation and fully characterized by gas chromatography (GC-FID) and gas chromatography-mass spectrometry (GC-MS). A total of 34 components (95.8%) were successfully identified in the essential oil which were characterized by high proportions of β-eudesmol (28.5%), caryophyllene oxide (9.5%), β-caryophyllene (7.2%), α-eudesmol (6.5%) and germacrene D (6.2%). Keywords: Ebenaceae, Diospyros wallichii, essential oil, hydrodistillation, β-eudesmol, GC-MS Introduction Essential oils are complex mixtures of volatile compounds, mainly terpenes and oxygenated aromatic and aliphatic compounds, such as phenols, alcohols, aldehydes, ketones, esters, ethers, and oxides, biosynthesized and accumulated in many plants (Dhifi et al., 2016). These naturally occurring mixtures of volatile compounds have been gaining increasing interest because of their wide range of applications in pharmaceutical, sanitary, cosmetics, perfume, food, and agricultural industries (Jugreet et al., 2020). The Ebenaceae family contains approximately 5 genera and 500 species. -

Diospyros Kaki) Chips: Instrumental, Sensory, and Consumer Input for Product Development
foods Article Texture of Hot-Air-Dried Persimmon (Diospyros kaki) Chips: Instrumental, Sensory, and Consumer Input for Product Development Rebecca R. Milczarek 1,* , Rachelle D. Woods 1, Sean I. LaFond 2, Jenny L. Smith 3, Ivana Sedej 1 , Carl W. Olsen 1, Ana M. Vilches 1, Andrew P. Breksa 1 and John E. Preece 3 1 United States Department of Agriculture—Agricultural Research Service, Western Regional Research Center, Healthy Processed Foods Research Unit, 800 Buchanan Street, Albany, CA 94710, USA; [email protected] (R.D.W.); [email protected] (I.S.); [email protected] (C.W.O.); [email protected] (A.M.V.); [email protected] (A.P.B.) 2 Department of Food Science & Technology, University of California, One Shields Avenue, Davis, CA 95616, USA; [email protected] 3 United States Department of Agriculture—Agricultural Research Service, National Clonal Germplasm Repository, One Shields Avenue, Davis, CA 95616, USA; [email protected] (J.L.S.); [email protected] (J.E.P.) * Correspondence: [email protected]; Tel.: +1-510-559-5656 Received: 10 September 2020; Accepted: 6 October 2020; Published: 10 October 2020 Abstract: Persimmon (Diospyros kaki) is an underutilized tree fruit. Previous studies have shown the feasibility of making a hot-air-dried, chip-style product from persimmon. However, the texture of this type of product has not been explored or connected to consumer preference. Thus, for dried samples representing 37 cultivars, this study aimed to (1) predict trained sensory panel texture attributes from instrumental measurements, (2) predict consumer liking from instrumental measurements and sensory texture attributes, and (3) elucidate whether astringency type affects dried product texture. -

Notes on Persimmons, Kakis, Date Plums, and Chapotes by STEPHEN A
Notes on Persimmons, Kakis, Date Plums, and Chapotes by STEPHEN A. SPONGBERG The genus Diospyros is not at present an important genus of orna- mental woody plants in North America, and while native persimmons once were valuable fruits in the eastern United States, the fruits pro- duced by Diospyros species no longer are important food items in the American home. In the countries of eastern Asia at least two species of Diospyros are among the most common trees encountered in door- yard gardens and orchards, where they are cultivated for their edible fruits as well as for other uses and for their ornamental beauty. J. J. Rein, a German traveler and author, wrote in 1889 that Diospyros kaki Linnaeus f. was "undeniably the most widely distributed, most important, and most beautiful fruit-tree in Japan, Corea, and North- ern China." And in Japan, where D. kaki is second in importance as an orchard crop only to citrus fruit, the kaki often is referred to as the national fruit (Childers, 1972). The rarity with which species of Diospyros are found in cultivation in cool-temperate North America is partially due to the fact that most are native to regions of tropical and subtropical climate and are not hardy in areas of temperate climate. A member of the Ebenaceae or Ebony Family, the genus contains upwards of 400 species that occur Stephen A. Spongberg is a horticultural taxonomist at the Arnold Arbore- tum. He participated in the Arboretum’s collecting trip to Japan and Korea in the fall of 1977, an experience which intensifted his interest in persim- mons. -

Medical and Cosmetic Applications of Persimmon (Diospyros Kaki L.) and Their Toxicity Assessment-A Review
Volume: 1 Issue: 3 International Journal of Traditional and Complementary Publisher Year: 2020 Medicine Research Duzce University REVIEW Medical and Cosmetic Applications of Persimmon (Diospyros kaki L.) and Their Toxicity Assessment-A review Ayse Kurt1* Ertugrul Kaya2 1 Traditional and Complementary Medicine Application and Research Center, Duzce University, Duzce, Turkey 2 Department of Pharmacology, Medicine Faculty, Duzce University, Duzce, Turkey *Corresponding Author: Ayse Kurt, e-mail: [email protected] Received: 27.07.2020 Accepted: 10.11.2020 Abstract In this review study, it is aimed to summarize the information cited about medical and cosmetic applications of the date persimmon (Diospyros kaki L.f.) and accordingly the toxicity assessment. For this purpose, the information cited about medical and cosmetic applications of the date persimmon (Diospyros kaki) and accordingly the toxicity assessment were summarized. Persimmon (Diospyros kaki), which is cultivated in tropical/subtropic regions such as China, Korea, Japan and Brazil, especially in the Far Eastern countries with more hot climate conditions, has been named as Trabzon Persimmon because it entered Turkey through the Black Sea region. This fruit type has a very important role on the immune system thanks to vitamins and some active ingredients. Although it is mostly consumed as fresh fruit and dried in our country, there are also formulations developed as medical support products (dematological and cosmetic applications etc.) in different countries worldwide. However, toxicity assessment studies on natural plants/herbal products are also very few. Since there is scientific evidence on the phytotherapeutic effects of Diospyros kaki, and the product scale on the market is very narrow, this is highly promising for future healthcare products. -

Rootstocks for the Oriental Persimmon
California Avocado association 1940 Yearbook 25: 43-44 Rootstocks for the Oriental Persimmon Robert W. Hodgson University of California, Los Angeles, Calif, From Proc. Amer. Soc. Hort. Sci. Vol. 37, 1939. Prior to 1919, when the federal plant quarantine went into effect, most of the Oriental persimmon (Diospyros kaki) trees planted in California were imported from Japan and were on the kaki rootstock. While extensive planting of this fruit did not occur until after that period, many of the old trees still remain in a condition of good vigor and a few are known which are now between 50 and 60 years old. Though at least a dozen varieties were introduced in these importations, no evidence of a rootstock problem seems to have been reported. For the past two decades, however, the only rootstock employed commercially has been the lotus persimmon (Diospyros lotus). It is not altogether clear why California nurserymen chose this rootstock, though the difficulty of obtaining seed of the Oriental persimmon was probably an important reason. For the Hachiya variety, which has comprised probably 98 per cent of the trees propagated, the results appear to be satisfactory thus far, though the characteristic vigor of the young trees seems to be associated with excessive shedding of the immature fruits. However, it early became evident that Fuyu, a non-astringent variety of Japanese origin, did not behave like Hachiya on this rootstock. The graft-union was usually poor, and most of the trees grew slowly, came into bearing early, and declined within a few years. Although some thousands of trees have been propagated, it is difficult to find good trees more than 10 years old, and the general experience has been that most of the trees have declined before attaining that age. -

Protective Effect of Diospyros Kaki Against Glucose-Oxygen-Serum Deprivation-Induced PC12 Cells Injury
Hindawi Publishing Corporation Advances in Pharmacological Sciences Volume 2016, Article ID 3073078, 5 pages http://dx.doi.org/10.1155/2016/3073078 Research Article Protective Effect of Diospyros kaki against Glucose-Oxygen-Serum Deprivation-Induced PC12 Cells Injury Fatemeh Forouzanfar,1 Shaghayegh Torabi,2 Vahid R. Askari,3 Elham Asadpour,4 and Hamid R. Sadeghnia1,2,3 1 Neurocognitive Research Center, School of Medicine, Mashhad University of Medical Sciences, Mashhad 917794-8564, Iran 2Pharmacological Research Center of Medicinal Plants, School of Medicine, Mashhad University of Medical Sciences, Mashhad917794-8564,Iran 3Department of Pharmacology, School of Medicine, Mashhad University of Medical Sciences, Mashhad 917794-8564, Iran 4Anesthesiology and Critical Care Research Center, Shiraz University of Medical Sciences, Shiraz, Iran Correspondence should be addressed to Hamid R. Sadeghnia; [email protected] Received 21 November 2015; Accepted 10 January 2016 Academic Editor: Masahiro Oike Copyright © 2016 Fatemeh Forouzanfar et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Ischemic cerebrovascular disease is one of the most common causes of death in the world. Recent interests have been focused on natural antioxidants and anti-inflammatory agents as potentially useful neuroprotective agents. Diospyros kaki (persimmon) has been shown to exert anti-inflammatory, antioxidant, and antineoplastic effects. However, its effects on ischemic damage have not been evaluated. Here, we used an in vitro model of cerebral ischemia and studied the effects of hydroalcoholic extract of peel (PeHE) and fruit pulp (PuHE) of persimmon on cell viability and markers of oxidative damage mainly intracellular reactive oxygen species (ROS) induced by glucose-oxygen-serum deprivation (GOSD) in PC12 cells. -

Diospyros Kaki (Japanese Persimmon) Size/Shape
Diospyros kaki (Japanese Persimmon) Asian persimmon is a deciduous tree grown for its late fall peach sized fruit. The tree has a beautiful glossy large leaf hairy on the bottom. The leaves turn orange or bonze in color during fall. Persimmons are commonly dioecious, female and male flowers are growing on different tree, but some cultivates like have both. As for the fruits there are two basic varieties, one which needs chill for the bitter taste to disappear , these fruits are mature after the leaves drop, the other new varieties matures early, no astringency taste from the beginning. Grows fruits best in full sun with good drainage, it tolerates a wide range of soils. Landscape Information French Name: Plaqueminier ﻛﺎﻛﻲ :Arabic Name Pronounciation: dye-OSS-pih-ross KACK-eye Plant Type: Tree Origin: China Heat Zones: 7, 8, 9, 10 Hardiness Zones: 6, 7, 8, 9 Uses: Topiary, Bonsai, Espalier, Specimen, Container, Edible Size/Shape Tree Shape: Upright Canopy Symmetry: Symmetrical Canopy Density: Dense Canopy Texture: Coarse Height at Maturity: 5 to 8 m Spread at Maturity: 3 to 5 meters Time to Ultimate Height: 10 to 20 Years Plant Image Diospyros kaki (Japanese Persimmon) Botanical Description Foliage Leaf Arrangement: Alternate Leaf Venation: Pinnate Leaf Persistance: Deciduous Leaf Type: Simple Leaf Blade: 5 - 10 cm Leaf Shape: Oval Leaf Margins: Undulate, Serrate Leaf Textures: Hairy, Glossy, Coarse Leaf Scent: No Fragance Color(growing season): Green Color(changing season): Yellow, Orange, Flower Image Copper Flower Flower Showiness: False -

Persimmon Postharvest Quality Maintenance Guidelines Carlos H
Persimmon 8/27/99 Postharvest Quality Maintenance Guidelines Carlos H. Crisosto Pomology Department University of California Davis, CA 95616 Scientific Name and Introduction Persimmon is usually the fruit of the species Diospyros kaki, a tree belonging to the family Ebenaceae and native to the Far East. It was originally cultivated in China and Japan, and it is therefore also known as the CHINESE DATE PLUM. Today persimmon is cultivated throughout the warmer parts of the world, e.g., in the South of France and other Mediterranean countries, as well as in the USA. The fruit is a berry resembling a tomato in color, shape and size, but the seeds are large, almond-shaped and few in number. The epidermis is thin, as in the tomato, and an enlarged calyx adheres to the base of the fruit. Persimmon has a delicious flavor and may be eaten as a dessert fruit, or it may be consumed dried or candied. In the USA a native species, Diospyros virginiana, occurs, but its fruit is very inferior to D. kaki. Another well known species is D. lotus, yielding DATE PLUMS and cultivated in the Orient and Italy. Quality Characteristics and Criteria Skin color from yellow to orange color with medium to large in size. Fruit should be firm (penetration force, using an 8-mm tip). It is suggested above 5 lb-force for 'Fuyu' and similar cultivars. Fruit should be freedom from growth cracks, mechanical injuries and decay. Soluble solids of 21-23% in 'Hachiya' and 18-20% in 'Fuyu' and similar non-astringent cultivars are recommended with no astringency (tannin content). -

Diospyros Kaki: Japanese Persimmon1 Edward F
ENH388 Diospyros kaki: Japanese Persimmon1 Edward F. Gilman and Dennis G. Watson2 Introduction General Information Japanese persimmon is a species related to common Scientific name: Diospyros kaki persimmon (Diospyros virginiana), but is native to Asia Pronunciation: dye-OSS-pih-ross KACK-eye (China, Japan). It can grow to about 30 feet when mature. Common name(s): Japanese persimmon This is an excellent fruit tree for ornamental use and makes Family: Ebenaceae an excellent specimen. The tree is a sight to behold when USDA hardiness zones: 7A through 9A (Fig. 2) leaves have fallen in autumn, displaying the bright yellow- Origin: not native to North America orange fruits throughout the canopy. Similar to common Invasive potential: little invasive potential persimmon, its preference is for a moist, well-drained soil Uses: container or planter; fruit; specimen in full sun locations. The tree has good drought tolerance. Availability: somewhat available, may have to go out of the Japanese persimmon develops an attractive red fall color, region to find the tree but the 2- to 4-inch-diameter fruits can be a big mess when they fall from the tree. Figure 2. Range Description Height: 20 to 30 feet Figure 1. Young Diospyros kaki: Japanese persimmon Spread: 15 to 25 feet Credits: Ed Gilman, UF/IFAS Crown uniformity: symmetrical 1. This document is ENH388, one of a series of the Environmental Horticulture Department, UF/IFAS Extension. Original publication date November 1993. Reviewed May 2014. Visit the EDIS website at http://edis.ifas.ufl.edu. 2. Edward F. Gilman, professor, Environmental Horticulture Department; Dennis G. -

Molecular Assisted Selection for Pollination-Constant and Non-Astringent Type Without Male Flowers in Spanish Germplasm for Persimmon Breeding
agronomy Article Molecular Assisted Selection for Pollination-Constant and Non-Astringent Type without Male Flowers in Spanish Germplasm for Persimmon Breeding Manuel Blasco 1, Francisco Gil-Muñoz 2, María del Mar Naval 1 and María Luisa Badenes 2,* 1 CANSO, Mestre Serrano, 1, 46250 L’Alcúdia, Valencia, Spain; [email protected] (M.B.); [email protected] (M.d.M.N.) 2 Instituto Valenciano de Investigaciones Agrarias, CV 315 km 10,5., 46113 Moncada, Valencia, Spain; [email protected] * Correspondence: [email protected] Received: 14 July 2020; Accepted: 7 August 2020; Published: 11 August 2020 Abstract: Persimmon (Diospyros kaki Thunb) species is a hexaploid genotype that has a morphologically polygamous gyonodioecious sexual system. D. kaki bears unisexual flowers. The presence of male flowers resulted in the presence of seeds in the varieties. The fruits of persimmon are classified according to their astringency and the pollination events that produced seeds and modify the levels of astringency in the fruit. The presence of seeds in astringent varieties as pollination variant astringent (PVA), pollination variant non-astringent (PVNA) and pollination constant astringent (PCA) resulted in fruits not marketable. Molecular markers that allow selection of the varieties according to the type of flowers at the plantlet stage would allow selection of seedless varieties. In this study, a marker developed in D. lotus by bulk segregant analysis (BSA) and amplified fragment length polymorphism (AFLP) markers, named DlSx-AF4, has been validated in a germplasm collection of persimmon, results obtained agree with the phenotype data. A second important trait in persimmon is the presence of astringency in ripened fruits.