Library: RCLIN6 Version: 6.20 Method: RCLIN6
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Legionella Shows a Diverse Secondary Metabolism Dependent on a Broad Spectrum Sfp-Type Phosphopantetheinyl Transferase
Legionella shows a diverse secondary metabolism dependent on a broad spectrum Sfp-type phosphopantetheinyl transferase Nicholas J. Tobias1, Tilman Ahrendt1, Ursula Schell2, Melissa Miltenberger1, Hubert Hilbi2,3 and Helge B. Bode1,4 1 Fachbereich Biowissenschaften, Merck Stiftungsprofessur fu¨r Molekulare Biotechnologie, Goethe Universita¨t, Frankfurt am Main, Germany 2 Max von Pettenkofer Institute, Ludwig-Maximilians-Universita¨tMu¨nchen, Munich, Germany 3 Institute of Medical Microbiology, University of Zu¨rich, Zu¨rich, Switzerland 4 Buchmann Institute for Molecular Life Sciences, Goethe Universita¨t, Frankfurt am Main, Germany ABSTRACT Several members of the genus Legionella cause Legionnaires’ disease, a potentially debilitating form of pneumonia. Studies frequently focus on the abundant number of virulence factors present in this genus. However, what is often overlooked is the role of secondary metabolites from Legionella. Following whole genome sequencing, we assembled and annotated the Legionella parisiensis DSM 19216 genome. Together with 14 other members of the Legionella, we performed comparative genomics and analysed the secondary metabolite potential of each strain. We found that Legionella contains a huge variety of biosynthetic gene clusters (BGCs) that are potentially making a significant number of novel natural products with undefined function. Surprisingly, only a single Sfp-like phosphopantetheinyl transferase is found in all Legionella strains analyzed that might be responsible for the activation of all carrier proteins in primary (fatty acid biosynthesis) and secondary metabolism (polyketide and non-ribosomal peptide synthesis). Using conserved active site motifs, we predict Submitted 29 June 2016 some novel compounds that are probably involved in cell-cell communication, Accepted 25 October 2016 Published 24 November 2016 differing to known communication systems. -

The Risk to Human Health from Free-Living Amoebae Interaction with Legionella in Drinking and Recycled Water Systems
THE RISK TO HUMAN HEALTH FROM FREE-LIVING AMOEBAE INTERACTION WITH LEGIONELLA IN DRINKING AND RECYCLED WATER SYSTEMS Dissertation submitted by JACQUELINE MARIE THOMAS BACHELOR OF SCIENCE (HONOURS) AND BACHELOR OF ARTS, UNSW In partial fulfillment of the requirements for the award of DOCTOR OF PHILOSOPHY in ENVIRONMENTAL ENGINEERING SCHOOL OF CIVIL AND ENVIRONMENTAL ENGINEERING FACULTY OF ENGINEERING MAY 2012 SUPERVISORS Professor Nicholas Ashbolt Office of Research and Development United States Environmental Protection Agency Cincinnati, Ohio USA and School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Professor Richard Stuetz School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Doctor Torsten Thomas School of Biotechnology and Biomolecular Sciences Faculty of Science The University of New South Wales Sydney, Australia ORIGINALITY STATEMENT '1 hereby declare that this submission is my own work and to the best of my knowledge it contains no materials previously published or written by another person, or substantial proportions of material which have been accepted for the award of any other degree or diploma at UNSW or any other educational institution, except where due acknowledgement is made in the thesis. Any contribution made to the research by others, with whom 1 have worked at UNSW or elsewhere, is explicitly acknowledged in the thesis. I also declare that the intellectual content of this thesis is the product of my own work, except to the extent that assistance from others in the project's design and conception or in style, presentation and linguistic expression is acknowledged.' Signed ~ ............................ -

Mycobacterium Tuberculosis: Assessing Your Laboratory
A more recent version of this document exists. View the 2019 Edition. Mycobacterium tuberculosis: Assessing Your Laboratory APHL Tool 2013 EDITION The following individuals contributed to the preparation of this edition of Mycobacterium tuberculosis: Assessing Your Laboratory Phyllis Della-Latta, PhD Columbia Presbyterian Medical Center Loretta Gjeltena, MA, MT(ASCP) National Laboratory Training Network Kenneth Jost, Jr. Texas Department of State Health Services Beverly Metchock, DrPH Centers for Disease Control and Prevention Glenn D. Roberts, PhD Mayo Clinic Max Salfinger, MD Florida Department of Health, Florida Bureau of Laboratories Dale Schwab, PhD, D(ABMM) Quest Diagnostics Julie Tans-Kersten Wisconsin State Laboratory of Hygiene Anthony Tran, MPH, MT(ASCP) Association of Public Health Laboratories David Warshauer, PhD, D(ABMM) Wisconsin State Laboratory of Hygiene Gail Woods, MD University of Texas Medical Branch Kelly Wroblewski, MPH, MT(ASCP) Association of Public Health Laboratories This publication was supported by Cooperative Agreement Number #1U60HM000803 between the Centers for Disease Control and Prevention (CDC) and the Association of Public Health Laboratories (APHL). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of CDC. © Copyright 2013, Association of Public Health Laboratories. All Rights Reserved. Table of Contents 1.0 Introduction ...................................................4 Background ...................................................4 Intended -

Biosynthesis of Isonitrile Lipopeptides by Conserved Nonribosomal Peptide Synthetase Gene Clusters in Actinobacteria
Biosynthesis of isonitrile lipopeptides by conserved nonribosomal peptide synthetase gene clusters in Actinobacteria Nicholas C. Harrisa, Michio Satob, Nicolaus A. Hermanc, Frederick Twiggc, Wenlong Caic, Joyce Liud, Xuejun Zhuc, Jordan Downeyc, Ryan Khalafe, Joelle Martine, Hiroyuki Koshinof, and Wenjun Zhangc,g,1 aDepartment of Plant and Microbial Biology, University of California, Berkeley, CA 94720; bDepartment of Pharmaceutical Sciences, University of Shizuoka, Shizuoka 422-8526, Japan; cDepartment of Chemical and Biomolecular Engineering, University of California, Berkeley, CA 94720; dDepartment of Bioengineering, University of California, Berkeley, CA 94720; eDepartment of Chemistry, University of California, Berkeley, CA 94720; fRIKEN Physical Center for Sustainable Resource Science, Wako, Saitama 3510198, Japan; and gChan Zuckerberg Biohub, San Francisco, CA 94158 Edited by Jerrold Meinwald, Cornell University, Ithaca, NY, and approved May 26, 2017 (received for review March 27, 2017) A putative lipopeptide biosynthetic gene cluster is conserved in many dependent oxidase, a fatty acyl-CoA thioesterase, an acyl-acyl species of Actinobacteria, including Mycobacterium tuberculosis and carrier protein ligase (AAL), an acyl carrier protein (ACP), and M. marinum, but the specific function of the encoding proteins has a single- or dimodule NRPS, respectively (Fig. 1 and SI Appendix, been elusive. Using both in vivo heterologous reconstitution and Fig. S1). Although all of these five proteins are typically involved in in vitro biochemical analyses, we have revealed that the five encod- secondary metabolite biosynthesis, the identity of the correspond- ing biosynthetic enzymes are capable of synthesizing a family of ing metabolite and the specific function of these proteins have not isonitrile lipopeptides (INLPs) through a thio-template mechanism. -

Is a Novel Proteasome Interactor in Mycobacteria and Related
RESEARCH ARTICLE Cdc48-like protein of actinobacteria (Cpa) is a novel proteasome interactor in mycobacteria and related organisms Michal Ziemski1, Ahmad Jomaa1, Daniel Mayer2, Sonja Rutz1, Christoph Giese1, Dmitry Veprintsev2†, Eilika Weber-Ban1* 1Institute of Molecular Biology & Biophysics, ETH Zurich, Zurich, Switzerland; 2Laboratory of Biomolecular Research, Paul Scherrer Institute, ETH Zurich, Villigen, Switzerland Abstract Cdc48 is a AAA+ ATPase that plays an essential role for many cellular processes in eukaryotic cells. An archaeal homologue of this highly conserved enzyme was shown to directly interact with the 20S proteasome. Here, we analyze the occurrence and phylogeny of a Cdc48 homologue in Actinobacteria and assess its cellular function and possible interaction with the bacterial proteasome. Our data demonstrate that Cdc48-like protein of actinobacteria (Cpa) forms hexameric rings and that the oligomeric state correlates directly with the ATPase activity. Furthermore, we show that the assembled Cpa rings can physically interact with the 20S core particle. Comparison of the Mycobacterium smegmatis wild-type with a cpa knockout strain under carbon starvation uncovers significant changes in the levels of around 500 proteins. Pathway mapping of the observed pattern of changes identifies ribosomal proteins as a particular hotspot, *For correspondence: [email protected] pointing amongst others toward a role of Cpa in ribosome adaptation during starvation. DOI: https://doi.org/10.7554/eLife.34055.001 Present address: †Centre of Membrane Proteins and Receptors, University of Birmingham and University of Introduction Nottingham, Nottingham, United Kingdom Energy-dependent chaperones and chaperone-protease complexes comprise important cellular components guarding protein homeostasis in all kingdoms of life. -

EVALUATION of the P45 MOBILE INTEGRATIVE ELEMENT and ITS ROLE IN
EVALUATION OF THE p45 MOBILE INTEGRATIVE ELEMENT AND ITS ROLE IN Legionella pneumophila VIRULENCE A Dissertation by LANETTE M. CHRISTENSEN Submitted to the Office of Graduate and Professional Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Chair of Committee, Jeffrey D. Cirillo Committee Members, James Samuel Jon Skare Farida Sohrabji Head of Program, Warren Zimmer May 2018 Major Subject: Medical Sciences Copyright 2018 Lanette Christensen ABSTRACT Legionella pneumophila are aqueous environmental bacilli that live within protozoal species and cause a potentially fatal form of pneumonia called Legionnaires’ disease. Not all L. pneumophila strains have the same capacity to cause disease in humans. The majority of strains that cause clinically relevant Legionnaires’ disease harbor the p45 mobile integrative genomic element. Contribution of the p45 element to L. pneumophila virulence and ability to withstand environmental stress were addressed in this study. The L. pneumophila Philadelphia-1 (Phil-1) mobile integrative element, p45, was transferred into the attenuated strain Lp01 via conjugation, designating p45 an integrative conjugative element (ICE). The resulting trans-conjugate, Lp01+p45, was compared with strains Phil-1 and Lp01 to assess p45 in virulence using a guinea pig model infected via aerosol. The p45 element partially recovered the loss of virulence in Lp01 compared to that of Phil-1 evident in morbidity, mortality, and bacterial burden in the lungs at the time of death. This phenotype was accompanied by enhanced expression of type II interferon in the lungs and spleens 48 hours after infection, independent of bacterial burden. -
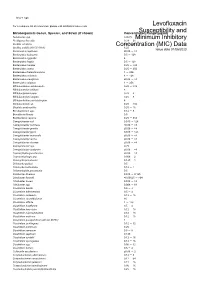
Susceptibility and Resistance Data
toku-e logo For a complete list of references, please visit antibiotics.toku-e.com Levofloxacin Microorganism Genus, Species, and Strain (if shown) Concentration Range (μg/ml)Susceptibility and Aeromonas spp. 0.0625 Minimum Inhibitory Alcaligenes faecalis 0.39 - 25 Bacillus circulans Concentration0.25 - 8 (MIC) Data Bacillus subtilis (ATCC 6051) 6.25 Issue date 01/06/2020 Bacteroides capillosus ≤0.06 - >8 Bacteroides distasonis 0.5 - 128 Bacteroides eggerthii 4 Bacteroides fragilis 0.5 - 128 Bacteroides merdae 0.25 - >32 Bacteroides ovatus 0.25 - 256 Bacteroides thetaiotaomicron 1 - 256 Bacteroides uniformis 4 - 128 Bacteroides ureolyticus ≤0.06 - >8 Bacteroides vulgatus 1 - 256 Bifidobacterium adolescentis 0.25 - >32 Bifidobacterium bifidum 8 Bifidobacterium breve 0.25 - 8 Bifidobacterium longum 0.25 - 8 Bifidobacterium pseudolongum 8 Bifidobacterium sp. 0.25 - >32 Bilophila wadsworthia 0.25 - 16 Brevibacterium spp. 0.12 - 8 Brucella melitensis 0.5 Burkholderia cepacia 0.25 - 512 Campylobacter coli 0.015 - 128 Campylobacter concisus ≤0.06 - >8 Campylobacter gracilis ≤0.06 - >8 Campylobacter jejuni 0.015 - 128 Campylobacter mucosalis ≤0.06 - >8 Campylobacter rectus ≤0.06 - >8 Campylobacter showae ≤0.06 - >8 Campylobacter spp. 0.25 Campylobacter sputorum ≤0.06 - >8 Capnocytophaga ochracea ≤0.06 - >8 Capnocytophaga spp. 0.006 - 2 Chlamydia pneumonia 0.125 - 1 Chlamydia psittaci 0.5 Chlamydia trachomatis 0.12 - 1 Chlamydophila pneumonia 0.5 Citrobacter diversus 0.015 - 0.125 Citrobacter freundii ≤0.00625 - >64 Citrobacter koseri 0.015 - -

Aquascreen® Legionella Species Qpcr Detection Kit
AquaScreen® Legionella species qPCR Detection Kit INSTRUCTIONS FOR USE FOR USE IN RESEARCH AND QUALITY CONTROL Symbols Lot No. Cat. No. Expiry date Storage temperature Number of reactions Manufacturer INDICATION The AquaScreen® Legionella species qPCR Detection kit is specifically designed for the quantitative detection of several Legionella species in water samples prepared with the AquaScreen® FastExt- ract kit. Its design complies with the requirements of AFNOR T90-471 and ISO/TS 12869:2012. Legionella are ubiquitous bacteria in surface water and moist soil, where they parasitize protozoa. The optimal growth temperature lies between +15 and +45 °C, whereas these gram-negative bacteria are dormant below 20 °C and do not survive above 60 °C. Importantly, Legionella are well-known as opportunistic intracellular human pathogens causing Legionnaires’ disease and Pontiac fever. The transmission occurs through inhalation of contami- nated aerosols generated by an infected source (e.g. human-made water systems like shower- heads, sink faucets, heaters, cooling towers, and many more). In order to efficiently prevent Legionella outbreaks, water safety control measures need syste- matic application but also reliable validation by fast Legionella testing. TEST PRINCIPLE The AquaScreen® Legionella species Kit uses qPCR for quantitative detection of legionella in wa- ter samples. In contrast to more time-consuming culture-based methods, AquaScreen® assays need less than six hours including sample preparation and qPCR to reliably detect Legionella. Moreover, the AquaScreen® qPCR assay has proven excellent performance in terms of specificity and sensitivity: other bacterial genera remain undetected whereas linear quantification is obtai- ned up to 1 x 106 particles per sample, therefore requiring no material dilution. -

Mycobacterium Bovis Isolates with M. Tuberculosis Specific Characteristics
gene (6). Furthermore, MTBC isolates can be differentiat- Mycobacterium ed by large sequence polymorphisms or regions of differ- ence (RD), and according to their distribution in the bovis Isolates with genome, a new phylogenetic scenario for the different species of the MTBC has been suggested (7–9). The pres- M. tuberculosis ence or absence of particular deletions has been proposed as being discriminative, e.g., lack of TdB1 for M. tubercu- Specific losis or lack of RD12 for M. bovis. In routine diagnostics, the combination of phenotypic Characteristics characteristics and biochemical features is sufficient to dif- ferentiate clinical M. bovis isolates, and in general, the Tanja Kubica,* Rimma Agzamova,† results obtained are unambiguous. However, here we Abigail Wright,‡ Galimzhan Rakishev,† describe the characteristics of 8 strains of the MTBC that Sabine Rüsch-Gerdes,* and Stefan Niemann* showed an unusual combination of phenotypic and bio- Our study is the first report of exceptional chemical attributes of both M. bovis and M. tuberculosis. Mycobacterium bovis strains that have some characteris- Molecular analyses confirmed the strains as M. bovis, tics of M. tuberculosis. The strains were isolated from 8 which in part have phenotypic and biochemical properties patients living in Kazakhstan. While molecular markers of M. tuberculosis. were typical for M. bovis, growth characteristics and bio- chemical test results were intermediate between M. bovis The Study and M. tuberculosis. During a previous investigation of 179 drug-resistant isolates from Kazakhstan (10), we determined the presence ycobacterium bovis causes tuberculosis (TB) mainly of 8 strains showing monoresistance to pyrazinamide. M in cattle but has a broad host range and causes dis- Kazakhstan is the largest of the central Asian republics and ease similar to that caused by M. -

Zoonotic Tuberculosis in Mammals, Including Bovine and Caprine
Zoonotic Importance Several closely related bacteria in the Mycobacterium tuberculosis complex Tuberculosis in cause tuberculosis in mammals. Each organism is adapted to one or more hosts, but can also cause disease in other species. The two agents usually found in domestic Mammals, animals are M. bovis, which causes bovine tuberculosis, and M. caprae, which is adapted to goats but also circulates in some cattle herds. Both cause economic losses including in livestock from deaths, disease, lost productivity and trade restrictions. They can also affect other animals including pets, zoo animals and free-living wildlife. M. bovis Bovine and is reported to cause serious issues in some wildlife, such as lions (Panthera leo) in Caprine Africa or endangered Iberian lynx (Lynx pardinus). Three organisms that circulate in wildlife, M. pinnipedii, M. orygis and M. microti, are found occasionally in livestock, Tuberculosis pets and people. In the past, M. bovis was an important cause of tuberculosis in humans worldwide. It was especially common in children who drank unpasteurized milk. The Infections caused by advent of pasteurization, followed by the establishment of control programs in cattle, Mycobacterium bovis, have made clinical cases uncommon in many countries. Nevertheless, this disease is M. caprae, M. pinnipedii, still a concern: it remains an important zoonosis in some impoverished nations, while wildlife reservoirs can prevent complete eradication in developed countries. M. M. orygis and M. microti caprae has also emerged as an issue in some areas. This organism is now responsible for a significant percentage of the human tuberculosis cases in some European countries where M. bovis has been controlled. -

Cytopathology: Mycobacterium Avium Complex Infection with Non-Necrotizing Granulomatous Inflammation Involving a Lymph Node
DEPARTMENT OF PATHOLOGY Case of the Week Cytopathology: Mycobacterium avium complex infection with non-necrotizing granulomatous inflammation involving a lymph node Prepared by: Andrea Hernandez, DO (resident), Dianne Grunes, MD (fellow), Barbara Bengston, CT, and Melissa Yee-Chang, DO (attending) February 16, 2016 History The patient is a 35 year old HIV-positive male who presents to the emergency department with altered mental status. He complained of low grade fever, productive cough and subjective weight loss. Oral candidiasis and painful cervical lymphadenopathy were noted on physical exam. His CD4 count was 40 cells/µL, indicating progression to AIDS. Chest X-ray demonstrated consolidation in the lower lobe of the left lung. An ultrasound guided aspiration biopsy of the cervical lymph node is performed. DC 2/4/2021 Figure. 1: (Diff-Quik stain, 400x magnification) Figure. 2: (Diff-Quik stain, 1000x magnification) DC 2/4/2021 Figure 3: (PAP stain, 40x magnification) Figure 1 - 3 Figure 1: Fine needle aspirate from the lymph node showing a histiocyte containing numerous outlines of intracellular bacilli within the cytoplasm. Figure 2: A histiocyte with abundant intracellular unstained bacilli which appear as slightly curved, colorless rods, displaying the “negative image” of the mycobacteria. Due to the striated appearance of the cellular cytoplasm, these histiocytes may be referred to as “pseudo-Gaucher cells”. Extracellular, negative-image mycobacteria are also seen within the background. Figure 3:Loose aggregate of epitheliod histiocytes forming a vague non-necrotizing granuloma, however, the mycobacteria are not readily identified as on the Diff Quik- stained smear. Diagnosis Mycobacterium avium complex infection with non-necrotizing granulomatous inflammation involving a lymph node Discussion Mycobacterium avium complex (MAC) infections are caused by one of two mycobacterial species: M. -

Mycobacterium Abscessus Pulmonary Disease: Individual Patient Data Meta-Analysis
ORIGINAL ARTICLE RESPIRATORY INFECTIONS Mycobacterium abscessus pulmonary disease: individual patient data meta-analysis Nakwon Kwak1, Margareth Pretti Dalcolmo2, Charles L. Daley3, Geoffrey Eather4, Regina Gayoso2, Naoki Hasegawa5, Byung Woo Jhun 6, Won-Jung Koh 6, Ho Namkoong7, Jimyung Park1, Rachel Thomson8, Jakko van Ingen 9, Sanne M.H. Zweijpfenning10 and Jae-Joon Yim1 @ERSpublications For Mycobacterium abscessus pulmonary disease in general, imipenem use is associated with improved outcome. For M. abscessus subsp. abscessus, the use of either azithromycin, amikacin or imipenem increases the likelihood of treatment success. http://ow.ly/w24n30nSakf Cite this article as: Kwak N, Dalcolmo MP, Daley CL, et al. Mycobacterium abscessus pulmonary disease: individual patient data meta-analysis. Eur Respir J 2019; 54: 1801991 [https://doi.org/10.1183/ 13993003.01991-2018]. ABSTRACT Treatment of Mycobacterium abscessus pulmonary disease (MAB-PD), caused by M. abscessus subsp. abscessus, M. abscessus subsp. massiliense or M. abscessus subsp. bolletii, is challenging. We conducted an individual patient data meta-analysis based on studies reporting treatment outcomes for MAB-PD to clarify treatment outcomes for MAB-PD and the impact of each drug on treatment outcomes. Treatment success was defined as culture conversion for ⩾12 months while on treatment or sustained culture conversion without relapse until the end of treatment. Among 14 eligible studies, datasets from eight studies were provided and a total of 303 patients with MAB-PD were included in the analysis. The treatment success rate across all patients with MAB-PD was 45.6%. The specific treatment success rates were 33.0% for M. abscessus subsp. abscessus and 56.7% for M.