Open Li Xingyan Thoracic.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diapositiva 1
Thoracic Cage and Thoracic Inlet Professor Dr. Mario Edgar Fernández. Parts of the body The Thorax Is the part of the trunk betwen the neck and abdomen. Commonly the term chest is used as a synonym for thorax, but it is incorrect. Consisting of the thoracic cavity, its contents, and the wall that surrounds it. The thoracic cavity is divided into 3 compartments: The central mediastinus. And the right and left pulmonary cavities. Thoracic Cage The thoracic skeleton forms the osteocartilaginous thoracic cage. Anterior view. Thoracic Cage Posterior view. Summary: 1. Bones of thoracic cage: (thoracic vertebrae, ribs, and sternum). 2. Joints of thoracic cage: (intervertebral joints, costovertebral joints, and sternocostal joints) 3. Movements of thoracic wall. 4. Thoracic cage. Thoracic apertures: (superior thoracic aperture or thoracic inlet, and inferior thoracic aperture). Goals of the classes Identify and describe the bones of the thoracic cage. Identify and describe the joints of thoracic cage. Describe de thoracic cage. Describe the thoracic inlet and identify the structures passing through. Vertebral Column or Spine 7 cervical. 12 thoracic. 5 lumbar. 5 sacral 3-4 coccygeal Vertebrae That bones are irregular, 33 in number, and received the names acording to the position which they occupy. The vertebrae in the upper 3 regions of spine are separate throughout the whole of life, but in sacral anda coccygeal regions are in the adult firmly united in 2 differents bones: sacrum and coccyx. Thoracic vertebrae Each vertebrae consist of 2 essential parts: An anterior solid segment: vertebral body. The arch is posterior an formed of 2 pedicles, 2 laminae supporting 7 processes, and surrounding a vertebral foramen. -
Sacral Dimple – Tethered Cord Pathway V3.0: Diagnosis
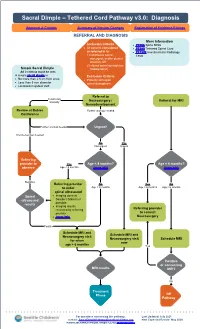
Sacral Dimple – Tethered Cord Pathway v3.0: Diagnosis Approval & Citation Summary of Version Changes Explanation of Evidence Ratings REFERRAL AND DIAGNOSIS More Information Inclusion Criteria • PE056 Spina Bifida • All patients considered • PE589 Tethered Spinal Cord or referred in for • PE1999 Anesthesia for Radiology 1) cutaneous sacral, Tests coccygeal, and/or gluteal anomaly OR 2) closed spinal dysraphism Simple Sacral Dimple (radiographic) All 3 criteria must be met. A simple sacral dimple is: Exclusion Criteria • No more than 2.5 cm from anus • Patients with open • Less than 5 mm diameter spinal dysraphism • Localized in gluteal cleft Referral to Cutaneous anomaly Neurosurgery / Referral for MRI Neurodevelopment Review at Babies Further workup needed Conference Further workup needed Urgent? No intervention needed No Yes Not urgent Urgent Referring provider to Yes Age < 4 months? Age < 6 months? observe Age < 4 months (more info) (more info) Negative Referring provider No Yes No to order Age ≥ 4 months Age < 6 months Age ≥ 6 months spinal ultrasound Spinal • Imaging done at ultrasound Seattle Children’s if results possible • Imaging results reviewed by referring Referring provider provider to consult • (more info) Neurosurgery Positive Schedule MRI and Schedule MRI and Neurosurgery visit Neurosurgery visit Schedule MRI for when now age > 6 months Yes Positive or concerning MRI results MRI? No Treatment Phase Off Pathway For questions concerning this pathway, Last Updated: July 2021 contact: [email protected] -

Anatomy of the Spine
12 Anatomy of the Spine Overview The spine is made of 33 individual bones stacked one on top of the other. Ligaments and muscles connect the bones together and keep them aligned. The spinal column provides the main support for your body, allowing you to stand upright, bend, and twist. Protected deep inside the bones, the spinal cord connects your body to the brain, allowing movement of your arms and legs. Strong muscles and bones, flexible tendons and ligaments, and sensitive nerves contribute to a healthy spine. Keeping your spine healthy is vital if you want to live an active life without back pain. Spinal curves When viewed from the side, an adult spine has a natural S-shaped curve. The neck (cervical) and low back (lumbar) regions have a slight concave curve, and the thoracic and sacral regions have a gentle convex curve (Fig. 1). The curves work like a coiled spring to absorb shock, maintain balance, and allow range of motion throughout the spinal column. The muscles and correct posture maintain the natural spinal curves. Good posture involves training your body to stand, walk, sit, and lie so that the least amount of strain is placed on the spine during movement or weight-bearing activities. Excess body weight, weak muscles, and other forces can pull at the spine’s alignment: • An abnormal curve of the lumbar spine is lordosis, also called sway back. • An abnormal curve of the thoracic spine is Figure 1. (left) The spine has three natural curves that form kyphosis, also called hunchback. an S-shape; strong muscles keep our spine in alignment. -

Vertebral Column
Vertebral Column • Backbone consists of Cervical 26 vertebrae. • Five vertebral regions – Cervical vertebrae (7) Thoracic in the neck. – Thoracic vertebrae (12) in the thorax. – Lumbar vertebrae (5) in the lower back. Lumbar – Sacrum (5, fused). – Coccyx (4, fused). Sacrum Coccyx Scoliosis Lordosis Kyphosis Atlas (C1) Posterior tubercle Vertebral foramen Tubercle for transverse ligament Superior articular facet Transverse Transverse process foramen Facet for dens Anterior tubercle • Atlas- ring of bone, superior facets for occipital condyles. – Nodding movement signifies “yes”. Axis (C2) Spinous process Lamina Vertebral foramen Transverse foramen Transverse process Superior articular facet Odontoid process (dens) •Axis- dens or odontoid process is body of atlas. – Pivotal movement signifies “no”. Typical Cervical Vertebra (C3-C7) • Smaller bodies • Larger spinal canal • Transverse processes –Shorter – Transverse foramen for vertebral artery • Spinous processes of C2 to C6 often bifid • 1st and 2nd cervical vertebrae are unique – Atlas & axis Typical Cervical Vertebra Spinous process (bifid) Lamina Vertebral foramen Inferior articular process Superior articular process Transverse foramen Pedicle Transverse process Body Thoracic Vertebrae (T1-T12) • Larger and stronger bodies • Longer transverse & spinous processes • Demifacets on body for head of rib • Facets on transverse processes (T1-T10) for tubercle of rib Thoracic Vertebra- superior view Spinous process Transverse process Facet for tubercle of rib Lamina Superior articular process -

The Spinal Canal in Cervical Spondylosis
J Neurol Neurosurg Psychiatry: first published as 10.1136/jnnp.26.2.166 on 1 April 1963. Downloaded from J. Neurol. Neurosurg. Psychiat., 1963, 26, 166 The spinal canal in cervical spondylosis A. R. CHRISPIN AND F. LEES From the Departments of Diagnostic Radiology and Neurology, St. Bartholomew's Hospital, London Cervical spondylosis was defined by Brain (1948), by the canal size with one of the patient's own para- Brain, Northfield, and Wilkinson (1952), and by meters in the region, namely, the size of the bodies of Frykholm (1951). Its essential features are protrusion the cervical vertebrae. This method eliminates geo- of the annulus fibrosus and changes in the adjacent metrical errors of radiographic projection and avoids vertebral margins. The size of the cervical spinal the difficulties in measuring the varying antero- canal may play a major part in the neurological posterior diameters of the irregular canal. manifestations of cervical spondylosis. The antero-posterior diameters of the cervical canal in 200 normal adults were measured by Boijsen METHOD (1954). Wolf, Khilnani, and Malis (1956) in a similar study found that the antero-posterior diameter at the The routinely taken lateral radiograph ofthecervicalspine first cervical vertebra varied 16 to be examined is placed on a horizontally positioned from mm. to more viewing box. A piece of blank developed x-ray film is than 30 mm. and from C.4 to C.7 varied from 12 mm. then placed over the radiograph. A careful tracing using Protected by copyright. to 22 mm. The average antero-posterior diameter a sharp film-marking pencil is then made. -

Spinal Stenosis.Pdf
Spinal Stenosis Overview Spinal stenosis is the narrowing of your spinal canal and nerve root canal along with the enlargement of your facet joints. Most commonly it is caused by osteoarthritis and your body's natural aging process, but it can also develop from injury or previous surgery. As the spinal canal narrows, there is less room for your nerves to branch out and move freely. As a result, they may become swollen and inflamed, which can cause pain, cramping, numbness or weakness in your legs, back, neck, or arms. Mild to moderate symptoms can be relieved with medications, physical therapy and spinal injections. Severe symptoms may require surgery. Anatomy of the spinal canal To understand spinal stenosis, it is helpful to understand how your spine works. Your spine is made of 24 moveable bones called vertebrae. The vertebrae are separated by discs, which act as shock absorbers preventing the vertebrae from rubbing together. Down the middle of each vertebra is a hollow space called the spinal canal that contains the spinal cord, spinal nerves, ligaments, fat, and blood vessels. Spinal nerves exit the spinal canal through the intervertebral foramen (also called the nerve root canal) to branch out to your body. Both the spinal and nerve root canals are surrounded by bone and ligaments. Bony changes can narrow the canals and restrict the spinal cord or nerves (see Anatomy of the Spine). What is spinal stenosis? Spinal stenosis is a degenerative condition that happens gradually over time and refers to: • narrowing of the spinal and nerve root canals • enlargement of the facet joints • stiffening of the ligaments • overgrowth of bone and bone spurs (Figure 1) Figure 1. -

Ultrasonographic Evaluation of the Sacrococcyx and Spinal Canal In
http:// ijp.mums.ac.ir Original Arti cle (Pages: 6267-6274) Ultrasonographic Evaluation of the Sacrococcyx and Spinal Canal in Children with Constipation Seyed Ali Alamdaran1, Mohammad Taghi Pourhoseini1, Seyed Ali Jafari2, Reza Shojaeian3, 1Ali Feyzi1, *Masoud Pezeshki Rad 1, 4 1 Department of Radiology, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran. 2 Department of Pediatrics, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran. 3 Department of Pediatric Surgery, Faculty of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran. 4 Surgical Oncology Research Center, Mashhad University of Medical Sciences, Mashhad, Iran. Abstract Background The occult neurological disorders are an important cause of constipation in children. This study aimed to evaluate the spinal canal and lumbosacral by ultrasound in pediatric constipation to better identifying neurological causes of constipation. Materials and Methods In this case-control study, 100 children with constipation (age range 1 to 14 years) without previously known chronic illness referred to the Radiology Department of the Mashhad Pediatric Dr. Sheikh Hospital were selected. After recording clinical data, the patients were undergone sacral and spinal cord ultrasound examinations and the results were compared with the control group (healthy children with transient illness (otherwise constipation or urinary disorders) who had referred to radiology department for sonographic examination). Results The mean age of patients was 6 ± 3.3 years old. The tethered cord and occult intrasacral meningocele were observed in 2% of patients group. Spina bifida was found in 64% patients, and 31% control subjects with significant difference (P = 0.009). About one third of cases with spina bifida were found in lumbar L5 vertebra, and another two third were in high sacral vertebrae (S1 or S2). -

C1 Stenosis – an Easily Missed Cause for Cervical Myelopathy
Neurospine 2019;16(3):456-461. Neurospine https://doi.org/10.14245/ns.1938200.100 pISSN 2586-6583 eISSN 2586-6591 Review Article C1 Stenosis – An Easily Missed Cause Corresponding Author for Cervical Myelopathy Andrei Fernandes Joaquim 1 2 3 2 https://orcid.org/0000-0003-2645-0483 Andrei Fernandes Joaquim , Griffin Baum , Lee A. Tan , K. Daniel Riew 1Neurosurgery Division, Department of Neurology, State University of Campinas (UNICAMP), Campinas-SP, Neurosurgery Division, Department of Brazil Neurology, State University of Campinas 2Department of Orthopedic Surgery, Columbia University, New York, NY, USA 3 (UNICAMP), Campinas, Brazil Department of Neurosurgery, University of California San Francisco, San Francisco, CA, USA E-mail: [email protected] Received: June 13, 2019 C1 stenosis is often an easily missed cause for cervical myelopathy. The vast majority of cer- Revised: July 16, 2019 vical myelopathy occurs in the subaxial cervical spine. The cervical canal is generally largest Accepted: July 17, 2019 at C1/2, explaining the relatively rare incidence of neurological deficits in patients with odon- toid fractures. However, some subjects have anatomical anomalies of the atlas, which may cause stenosis and result in clinical symptoms similar to subaxial cord compression. Isolat- ed pure atlas hypoplasia leading to stenosis is quite rare and may be associated with other This is an Open Access article distributed under anomalies, such as atlas clefts or transverse ligament calcification. It may also be more com- the terms of the Creative Commons Attribution monly associated with syndromic conditions such as Down or Turner syndrome. Although Non-Commercial License (http://creativecom- mons.org/licenses/by-nc/4.0/) which permits the diagnosis can be easily made with a cervical magnetic resonance imaging, the C3/2 spi- unrestricted non-commercial use, distribution, nolaminar test using a lateral cervical plain radiograph is a useful and sensitive tool for screen- and reproduction in any medium, provided the ing. -

Diagnosis and Treatment of Lumbar Spinal Canal Stenosis
Ⅵ Low Back Pains Diagnosis and Treatment of Lumbar Spinal Canal Stenosis JMAJ 46(10): 439–444, 2003 Katsuro TOMITA Department of Orthopedic Surgery, Kanazawa University Abstract: Lumbar spinal canal stenosis is a syndrome of neurological symptoms that appear due to compression of the cauda equina nerve bundle and nerve roots, as a result of narrowing of the lumbar spinal canal through which the spinal nerve bundle passes, and accompanies the degeneration that occurs with aging. Specific causes related to narrowing and compression are degenerative bulging of an intervertebral disk; thickening of a vertebral arch, an apophyseal joint or the yellow ligament; and spondylolisthesis. All these factors, which are due to various dis- eases, cause narrowing of the spinal canal, resulting in compression of the spinal nerves inside the canal and inducing neurological symptoms. The main symptoms are sciatica and intermittent claudication that are treated with therapies based on the severity of the stenosis. These range from conservative treatment provided at pain clinics etc. and rehabilitation, to surgical treatment. Especially in recent years, lumbar spinal canal stenosis has been treated increasingly in the elderly. Key words: Lumbar spine; Low back pain; Spinal canal stenosis; Intermittent claudication; Sciatica; Nerve root block What is Lumbar Spinal Canal Stenosis? vertebral arch, an apophyseal joint or the yel- low ligament; and spondylolisthesis. Lumbar spinal canal stenosis is a syndrome These factors, due to various diseases, cause of symptoms that appear due to compression of stenosis of the spinal canal, resulting in com- the cauda equina nerve bundle and nerve roots, pression of the spinal nerves inside the canal, as a result of narrowing of the lumbar spinal thus inducing neurological symptoms. -

Neurological Manifestation of Sacral Tumors
Neurosurg Focus 15 (2):Article 1, 2003, Click here to return to Table of Contents Neurological manifestation of sacral tumors MICHAEL PAYER, M.D. Department of Neurosurgery, University Hopital of Geneva, Switzerland An extensive analysis of the existing literature concerning sacral tumors was conducted to characterize their clin- ical manifestations. Although certain specific manifestations can be attributed to some of the tumor types, a more general pattern of clinical presentation of an expansive sacral lesion can be elaborated. Local pain with or without pseudoradicular or radicular radiation is the most frequent initial symptom and is usually followed by the manifesta- tion of a lumbosacral sensorimotor deficit; bladder/bowel and/or sexual dysfunction appear throughout the natural course of disease. KEY WORDS • sacrum • tumor • lesion • neurological presentation All sacral and presacral tumors are rare.32,93 In one se- REVIEW OF SACRAL ANATOMY ries patients with these tumors were estimated to account for approximately one in 40,000 hospital admissions.93 Osseous Structures of the Sacrum Tumors arising from the bone of the sacrum are by far the The sacrum is a complex bone, comprising five sacral most frequent sacral tumors; chordomas are the most com- vertebrae that have fused. In its center lies the longitudi- mon and GCTs the second most common.20,46,50,61,74,81,98 nal sacral canal, which opens caudally posteriorly into the Although sacrococcygeal teratoma is the most common sacral hiatus, an incomplete posterior closure of the S-5 sacral tumor in neonates, it is very rare in adults.30,45,66 lamina. The thick anterior or pelvic face of the sacrum is The author conducted an extensive analysis of the exist- concave and contains four right- and left-sided anterior ing literature concerning tumors of the sacrum to charac- sacral foramina. -

Congenital Narrowing of the Cervical Spinal Canal
J Neurol Neurosurg Psychiatry: first published as 10.1136/jnnp.38.12.1218 on 1 December 1975. Downloaded from Journal ofNeurology, Neurosurgery, and Psychiatry, 1975, 38, 1218-1224 Congenital narrowing of the cervical spinal canal JEFFREY T. KESSLER From the Department ofNeurology, The New York Hospital-Cornell Medical Center, New York, New York 10021, U.S.A. SYNOPSIS The clinical and laboratory findings in six patients with congenital narrowing of the cervical spinal canal and neurological symptoms are described. A variable age of onset and an entirely male occurrence were found. Signs and symptoms of spinal cord dysfunction predominated in all but one patient. Symptoms were produced in five patients by increased physical activity alone. Congenital narrowing of the cervical spinal canal may result in cord compression without a history of injury and occasionally without evidence of significant bony degenerative changes. The clinical features may be distinguishable from those found in cervical spondylosis without congenital nar- rowing. Intermittent claudication of the cervical spinal cord appears to be an important feature of this syndrome. Surgery improved four out of five people. guest. Protected by copyright. The cervical spinal canal is a triangular tube can be found in most large series of this disorder that diminishes in size from top to bottom (Payne and Spillane, 1957; Epstein et al., 1963; (Burrows, 1963). The transverse (interpedicular) Crandall and Batzdorf, 1966; Symon and Lav- diameter is nearly twice that of the sagittal ender, 1967; Galera and Tovi, 1968; Wilkinson (anteroposterior) diameter. Thus, the most et al., 1969; Eyssette et al., 1970; Godlewski, critical radiographic measurement in the cer- 1972; Nurick, 1972a). -

Age Changes in the Lumbar Spinal and Intervertebral Canals
Paraplegia 26 (1988) 238-249 1988 International Medical Society of Paraplegia Age Changes in the Lutnbar Spinal and Intervertebral Canals L. Twontey, BAppSc, BSc(Hons), PhD,l J. Taylor, MB, ehB, PhD2 lSchool of Physiotherapy, Curtin University of Technology, Selby Street, Shenton Park, Western Australia, 6008, 2Department of Anatomy and Human Biology, University of Western Australia, Stirling Highway, Crawley, Western Australia, 6009. Suntntary A variety of measurements of the lumbar spinal canal (SC) and intervertebral foramina (lVF) are presented and compared in two adult age groups (48 individuals aged 20-35 and 48 individuals over 60 years) in both males and females. The dimensions measured in fresh post-mortem material include: mid-sagittal (AP) and transverse diameters (TD) of the SC; height, length and greater antero-posterior (AP) width of the IVF and ligamentum flavum (LF) thickness at the lumbo-sacral level. The data show a significant decline in the SC APD in both sexes, but the SC TD increases with ageing in males while it declines with ageing in females. The IVF height decreases with ageing but its AP width increases and its length increases. The LF shows a 50�o increase in thickness with ageing. These ageing changes in a 'normal' population provide the neurovascular bundle in the IVF with less 'room for manoeuvre' and support the view that minor additional pathology is more likely to lead to entrapment syndromes in the elderly than in the young. Key words: Spinal canal; Intervertebral foramina; Lumbar spine; Spinal stenosis; Age changes. The spinal canal (SC) and the intervertebral foramina (IVF) of the lumbar spine serve as containers and pathways for the terminal 2 cm of the spinal cord, the cauda equina and the nerve roots of the lumbar spinal nerves (Schmorl and Junghanns, 1971).