Inverse Molecular Docking As a Novel Approach to Study Anticarcinogenic and Anti-Neuroinflammatory Effects of Curcumin
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Caractérisation De La Polarisation Des Macrophages Pulmonaires Humains Et Voies De Régulation Charlotte Abrial
Caractérisation de la polarisation des macrophages pulmonaires humains et voies de régulation Charlotte Abrial To cite this version: Charlotte Abrial. Caractérisation de la polarisation des macrophages pulmonaires humains et voies de régulation. Biologie cellulaire. Université de Versailles-Saint Quentin en Yvelines, 2014. Français. NNT : 2014VERS0033. tel-01326578 HAL Id: tel-01326578 https://tel.archives-ouvertes.fr/tel-01326578 Submitted on 8 Dec 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Université de Versailles Saint Quentin en Yvelines UFR DES SCIENCES DE LA SANTÉ École doctorale GAO "Des génomes aux organismes" Année universitaire 2014 – 2015 N° le 03 novembre 2014 THESE DE DOCTORAT Présentée pour l’obtention du grade de DOCTEUR DE L’UNIVERSITÉ VERSAILLES – SAINT QUENTIN EN YVELINES Spécialité : Biologie cellulaire Par Charlotte ABRIAL Caractérisation de la polarisation des macrophages pulmonaires humains et voies de régulation Composition du jury: Directeur de thèse Pr. DEVILLIER Philippe Rapporteur Dr. FROSSARD Nelly Rapporteur Pr. LAGENTE Vincent Examinateur Dr. TOUQUI Lhousseine Université de Versailles Saint Quentin en Yvelines UFR DES SCIENCES DE LA SANTÉ École doctorale GAO "Des génomes aux organismes" Année universitaire 2014 – 2015 N° THESE DE DOCTORAT Présentée pour l’obtention du grade de DOCTEUR DE L’UNIVERSITÉ VERSAILLES – SAINT QUENTIN EN YVELINES Spécialité : Biologie cellulaire Par Charlotte ABRIAL Caractérisation de la polarisation des macrophages pulmonaires humains et voies de régulation Composition du jury: Directeur de thèse Pr. -

Deutsche Gesellschaft Für Experimentelle Und Klinische Pharmakologie Und Toxikologie E.V
Naunyn-Schmiedeberg´s Arch Pharmacol (2013 ) 386 (Suppl 1):S1–S104 D OI 10.1007/s00210-013-0832-9 Deutsche Gesellschaft für Experimentelle und Klinische Pharmakologie und Toxikologie e.V. Abstracts of the 79 th Annual Meeting March 5 – 7, 2013 Halle/Saale, Germany This supplement was not sponsored by outside commercial interests. It was funded entirely by the publisher. 123 S2 S3 001 003 Multitarget approach in the treatment of gastroesophagel reflux disease – Nucleoside Diphosphate Kinase B is a Novel Receptor-independent Activator of comparison of a proton-pump inhibitor with STW 5 G-protein Signaling in Clinical and Experimental Atrial Fibrillation Abdel-Aziz H.1,2, Khayyal M. T.3, Kelber O.2, Weiser D.2, Ulrich-Merzenich G.4 Abu-Taha I.1, Voigt N.1, Nattel S.2, Wieland T.3, Dobrev D.1 1Inst. of Pharmaceutical & Medicinal Chemistry, University of Münster Pharmacology, 1Universität Duisburg-Essen Institut für Pharmakologie, Hufelandstr. 55, 45122 Essen, Hittorfstr 58-62, 48149 Münster, Germany Germany 2Steigerwald Arzneimittelwerk Wissenschaft, Havelstr 5, 64295 Darmstadt, Germany 2McGill University Montreal Heart Institute, 3655 Promenade Sir-William-Osler, Montréal 3Faculty of Pharmacy, Cairo University Pharmacology, Cairo Egypt Québec H3G 1Y6, Canada 4Medizinische Poliklinik, University of Bonn, Wilhelmstr. 35-37, 53111 Bonn, Germany 3Medizinische Fakultät Mannheim der Universität Heidelberg Institutes für Experimentelle und Klinische Pharmakologie und Toxikologie, Maybachstr. 14, 68169 Gastroesophageal reflux disease (GERD) was the most common GI-diagnosis (8.9 Mannheim, Germany million visits) in the US in 2012 (1). Proton pump inhibitors (PPI) are presently the mainstay of therapy, but in up to 40% of the patients complete symptom control fails. -

ARTICLE Doi: 10.12032/ATR20200603
ARTICLE doi: 10.12032/ATR20200603 Asian Toxicology Research A network pharmacology approach combined with animal experiment to investigate the blood enriching effect of Gei herba Wen-Bi Mu1, 2#, Can-Can Duan1, 2#, Zhi-Ping Zhong1, 2, Kuan Chen1, 2, Jian-Yong Zhang1, 2* 1School of Pharmacy, Zunyi Medical University, Zunyi 563000, China; 2Key Laboratory of Basic Pharmacology of Ministry of Education and Joint International Research Laboratory of Ethnomedicine of Ministry of Education, Zunyi Medical University, Zunyi 563000, China. #Authors contributed equally to this article. *Corresponding to: Jian-Yong Zhang. Zunyi Medical University, No. 6 Xuefu West Road, Xinpu District, Zunyi 563000, China. Email: [email protected]. Highlights (1) A network pharmacology approach and animal experiments were established to explore the nourishing blood effect of Lanbuzheng (Gei herba). (2) The main active components, targets and pathways of Lanbuzheng (Gei herba) of blood deficiency were predicted by network pharmacology. (3) It’s verified that Lanbuzheng (Gei herba) can treat blood deficiency by improving the peripheral blood routine index and organ index in animal experiment. Submit a manuscript: https://www.tmrjournals.com/atr ATR | August 2020 | vol. 2 | no. 3 | 109 doi: 10.12032/ATR20200603 ARTICLE Abstract Background: To explore active components of Lanbuzheng (Gei herba) and its underlying complex mechanism in treating blood deficiency induced by chemotherapy drug based on network pharmacology and mice experimental validation. Methods: Active components of Lanbuzheng (Gei herba) were screened by Lipinski’s rule of five. Targets acted with active components were predicted by PharmMapper database, and targets whose function associated with blood deficiency were screened by Therapeutic Target Database and UniProt. -

Untersuchungen Zur Regulation Der Polyphenolbiosynthese in Der Erdbeerfrucht (Fragaria Ananassa) Mittels Metabolite Profiling
TECHNISCHE UNIVERSITÄT MÜNCHEN Fachgebiet Biotechnologie der Naturstoffe Untersuchungen zur Regulation der Polyphenolbiosynthese in der Erdbeerfrucht (Fragaria ananassa) mittels Metabolite Profiling Ludwig F. M. Ring Vollständiger Abdruck der von der Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt der Technischen Universität München zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften genehmigten Dissertation. Vorsitzende: Univ.-Prof. Dr. B. Poppenberger Prüfer der Dissertation: 1. Univ.-Prof. Dr. W. Schwab 2. Univ.-Prof. Dr. Th. Hofmann 3. Univ.-Prof. Dr. D. R. Treutter Die Dissertation wurde am 17.06.2013 bei der Technischen Universität München eingereicht und durch die Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt am 22.10.2013 angenommen. „… and all the pieces matter“ Lester Freamon, 2002 meiner Familie Danksagung I Danksagung Meinem Doktorvater Prof. Dr. Wilfried Schwab gilt mein besonderer Dank für die Überlassung des Themas und die Möglichkeit an seinem Fachgebiet zu promovieren. Außerdem danke ich ihm für seine immerwährende Unterstützung und seinen ausstrahlenden Optimismus. Bei Prof. Dr. Brigitte Poppenberger, Prof. Dr. Thomas Hofmann und Prof. Dr. Dieter Treutter bedanke ich mich für die Mitarbeit in der Prüfungskommission. Allen Kooperationspartnern des FraGenomics-Projekts, insbesondere Prof. Dr. Juan Muñoz- Blanco, Dr. Beatrice Denoyes-Rothan und Dr. Amparo Monfort, möchte ich für die gute Zusammenarbeit und die fruchtbaren Diskussionen bei den Projekttreffen danken. Prof. Dr. Victoriano Valpuesta danke ich sehr für die Möglichkeit meine Arbeiten zur Proteinanalytik am Department für Molekularbiologie und Biochemie der Universität Málaga durchführen zu können. Seinem gesamten Arbeitskreis danke ich für die herzliche Aufnahme! Gracias a todos los miembros del grupo! Además, les agradesco a Dra. -

In Silico Prediction of the Mode of Action of Viola Odorata in Diabetes
Hindawi BioMed Research International Volume 2020, Article ID 2768403, 13 pages https://doi.org/10.1155/2020/2768403 Research Article In Silico Prediction of the Mode of Action of Viola odorata in Diabetes Manal Ali Buabeid ,1 El-Shaimaa A. Arafa ,1,2 Waseem Hassan ,3 and Ghulam Murtaza 3 1College of Pharmacy and Health Sciences, Ajman University, Ajman 346, UAE 2Department of Pharmacology and Toxicology, Faculty of Pharmacy, Beni-Suef University, Beni Suef 62514, Egypt 3Department of Pharmacy, COMSATS University Islamabad, Lahore Campus, 54000, Pakistan Correspondence should be addressed to Manal Ali Buabeid; [email protected] Received 16 April 2020; Revised 27 June 2020; Accepted 5 October 2020; Published 31 October 2020 Academic Editor: K. H. Mok Copyright © 2020 Manal Ali Buabeid et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. The metabolic syndrome increases the risk of different diseases such as type 2 diabetes. The prevalence of metabolic syndrome has rapidly grown and affected more than 230 million people worldwide. Viola odorata is a traditionally used plant for the treatment of diabetes; however, its mechanism to manage diabetes is still unknown. Purpose. This study was designed to systematically assess the mechanism of action of Viola odorata in diabetes. Methods. An extensive literature search was made to establish an ingredient-target database of Viola odorata. Of these, targets related to diabetes were identified and used to develop a protein-protein interaction network (PPIN) by utilizing the STITCH database. -
![Alternative Treatments for Cancer Prevention and Cure [Part 1]](https://docslib.b-cdn.net/cover/6616/alternative-treatments-for-cancer-prevention-and-cure-part-1-2356616.webp)
Alternative Treatments for Cancer Prevention and Cure [Part 1]
Advances in Pharmacology and Clinical Trials ISSN: 2474-9214 Alternative Treatments for Cancer Prevention and Cure [Part 1] Abdul Kader Mohiuddin* Review Article Secretary & Treasurer Dr M. Nasirullah Memorial Trust, Tejgaon, Dhaka, Bangladesh Volume 4 Issue 4 Received Date: September 02, 2019 *Corresponding author: Abdul Kader Mohiuddin, Secretary & Treasurer Dr M Published Date: October 17, 2019 Nasirullah Memorial Trust, Tejgaon, Dhaka, Bangladesh, Tel: +8802-9110553; Email: DOI: 10.23880/apct-16000168 [email protected] Abstract Many lay people along with some so called “key opinion leaders” have a common slogan “There's no answer for cancer”. Again, mistake delays proper treatment and make situation worse, more often. Compliance is crucial to obtain optimal health outcomes, such as cure or improvement in QoL. Patients may delay treatment or fail to seek care because of high out-of- pocket expenditures. Despite phenomenal development, conventional therapy falls short in cancer management. There are two major hurdles in anticancer drug development: dose-limiting toxic side effects that reduce either drug effectiveness or the QoL of patients and complicated drug development processes that are costly and time consuming. Cancer patients are increasingly seeking out alternative medicine and might be reluctant to disclose its use to their oncology treatment physicians. But there is limited available information on patterns of utilization and efficacy of alternative medicine for patients with cancer. As adjuvant therapy, many traditional medicines shown efficacy against brain, head and neck, skin, breast, liver, pancreas, kidney, bladder, prostate, colon and blood cancers. The literature reviews non-pharmacological interventions used against cancer, published trials, systematic reviews and meta-analyses. -

European Patent Office U.S. Patent and Trademark Office
EUROPEAN PATENT OFFICE U.S. PATENT AND TRADEMARK OFFICE CPC NOTICE OF CHANGES 89 DATE: JULY 1, 2015 PROJECT RP0098 The following classification changes will be effected by this Notice of Changes: Action Subclass Group(s) Symbols deleted: C12Y 101/01063 C12Y 101/01128 C12Y 101/01161 C12Y 102/0104 C12Y 102/03011 C12Y 103/01004 C12Y 103/0103 C12Y 103/01052 C12Y 103/99007 C12Y 103/9901 C12Y 103/99013 C12Y 103/99021 C12Y 105/99001 C12Y 105/99002 C12Y 113/11013 C12Y 113/12012 C12Y 114/15002 C12Y 114/99028 C12Y 204/01119 C12Y 402/01052 C12Y 402/01058 C12Y 402/0106 C12Y 402/01061 C12Y 601/01025 C12Y 603/02027 Symbols newly created: C12Y 101/01318 C12Y 101/01319 C12Y 101/0132 C12Y 101/01321 C12Y 101/01322 C12Y 101/01323 C12Y 101/01324 C12Y 101/01325 C12Y 101/01326 C12Y 101/01327 C12Y 101/01328 C12Y 101/01329 C12Y 101/0133 C12Y 101/01331 C12Y 101/01332 C12Y 101/01333 CPC Form – v.4 CPC NOTICE OF CHANGES 89 DATE: JULY 1, 2015 PROJECT RP0098 Action Subclass Group(s) C12Y 101/01334 C12Y 101/01335 C12Y 101/01336 C12Y 101/01337 C12Y 101/01338 C12Y 101/01339 C12Y 101/0134 C12Y 101/01341 C12Y 101/01342 C12Y 101/03043 C12Y 101/03044 C12Y 101/98003 C12Y 101/99038 C12Y 102/01083 C12Y 102/01084 C12Y 102/01085 C12Y 102/01086 C12Y 103/01092 C12Y 103/01093 C12Y 103/01094 C12Y 103/01095 C12Y 103/01096 C12Y 103/01097 C12Y 103/0701 C12Y 103/08003 C12Y 103/08004 C12Y 103/08005 C12Y 103/08006 C12Y 103/08007 C12Y 103/08008 C12Y 103/08009 C12Y 103/99032 C12Y 104/01023 C12Y 104/01024 C12Y 104/03024 C12Y 105/01043 C12Y 105/01044 C12Y 105/01045 C12Y 105/03019 C12Y 105/0302 -

The Plant Short-Chain Dehydrogenase (SDR) Superfamily: Genome-Wide Inventory and Diversification Patterns
Moummou et al. BMC Plant Biology 2012, 12:219 http://www.biomedcentral.com/1471-2229/12/219 RESEARCH ARTICLE Open Access The Plant Short-Chain Dehydrogenase (SDR) superfamily: genome-wide inventory and diversification patterns Hanane Moummou1,2, Yvonne Kallberg3, Libert Brice Tonfack1,4, Bengt Persson5,6 and Benoît van der Rest1,7* Abstract Background: Short-chain dehydrogenases/reductases (SDRs) form one of the largest and oldest NAD(P)(H) dependent oxidoreductase families. Despite a conserved ‘Rossmann-fold’ structure, members of the SDR superfamily exhibit low sequence similarities, which constituted a bottleneck in terms of identification. Recent classification methods, relying on hidden-Markov models (HMMs), improved identification and enabled the construction of a nomenclature. However, functional annotations of plant SDRs remain scarce. Results: Wide-scale analyses were performed on ten plant genomes. The combination of hidden Markov model (HMM) based analyses and similarity searches led to the construction of an exhaustive inventory of plant SDR. With 68 to 315 members found in each analysed genome, the inventory confirmed the over-representation of SDRs in plants compared to animals, fungi and prokaryotes. The plant SDRs were first classified into three major types —‘classical’, ‘extended’ and ‘divergent’—but a minority (10% of the predicted SDRs) could not be classified into these general types (‘unknown’ or ‘atypical’ types). In a second step, we could categorize the vast majority of land plant SDRs into a set of 49 families. Out of these 49 families, 35 appeared early during evolution since they are commonly found through all the Green Lineage. Yet, some SDR families — tropinone reductase-like proteins (SDR65C), ‘ABA2-like’-NAD dehydrogenase (SDR110C), ‘salutaridine/menthone-reductase-like’ proteins (SDR114C), ‘dihydroflavonol 4-reductase’-like proteins (SDR108E) and ‘isoflavone-reductase-like’ (SDR460A) proteins — have undergone significant functional diversification within vascular plants since they diverged from Bryophytes. -

Systems Pharmacological Approach to Investigate the Mechanism of Acori Tatarinowii Rhizoma for Alzheimer’S Disease
Hindawi Evidence-Based Complementary and Alternative Medicine Volume 2018, Article ID 5194016, 20 pages https://doi.org/10.1155/2018/5194016 Research Article Systems Pharmacological Approach to Investigate the Mechanism of Acori Tatarinowii Rhizoma for Alzheimer’s Disease Zhenyan Song , Fang Yin, Biao Xiang, Bin Lan, and Shaowu Cheng Te Key Laboratory of Hunan Province for Integrated Traditional Chinese and Western Medicine on Prevention and Treatment of Cardio-Cerebral Diseases, Hunan University of Chinese Medicine, Changsha, Hunan 410208, China Correspondence should be addressed to Shaowu Cheng; [email protected] Received 30 March 2018; Accepted 30 May 2018; Published 27 June 2018 Academic Editor: Ling Yang Copyright © 2018 Zhenyan Song et al. Tis is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. In traditional Chinese medicine (TCM), Acori Tatarinowii Rhizoma (ATR) is widely used to treat memory and cognition dysfunction. Tis study aimed to confrm evidence regarding the potential therapeutic efect of ATR on Alzheimer’s disease (AD) using a system network level based in silico approach. Study results showed that the compounds in ATR are highly connected to AD-related signaling pathways, biological processes, and organs. Tese fndings were confrmed by compound-target network, target-organ location network, gene ontology analysis, and KEGG pathway enrichment analysis. Most compounds in ATR have been reported to have antifbrillar amyloid plaques, anti-tau phosphorylation, and anti-infammatory efects. Our results indicated that compounds in ATR interact with multiple targets in a synergetic way. -

Molecular Biomarkers: Tools of Medicine
BioMed Research International Molecular Biomarkers: Tools of Medicine Guest Editors: Prabir K. Mandal, Shivani Soni, R. Renee Reams, Tiziano Verri, and Sudhish Mishra Molecular Biomarkers: Tools of Medicine BioMed Research International Molecular Biomarkers: Tools of Medicine Guest Editors: Prabir K. Mandal, Shivani Soni, R. Renee Reams, Tiziano Verri, and Sudhish Mishra Copyright © 2013 Hindawi Publishing Corporation. All rights reserved. This is a special issue published in “BioMed Research International.” All articles are open access articles distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Contents Molecular Biomarkers: Tools of Medicine, Prabir K. Mandal, Shivani Soni, R. Renee Reams, Tiziano Verri, Anita Mandal, and Sudhish Mishra Volume 2013, Article ID 595496, 2 pages Defects in Base Excision Repair Sensitize Cells to Manganese in S. cerevisiae, Adrienne P. Stephenson, Tryphon K. Mazu, Jana S. Miles, Miles D. Freeman, R. Renee Reams, and Hernan Flores-Rozas Volume 2013, Article ID 295635, 9 pages Practical Guidance for Implementing Predictive Biomarkers into Early Phase Clinical Studies, Matthew J. Marton and Russell Weiner Volume 2013, Article ID 891391, 9 pages Circulating microRNAs and Kallikreins before and after Radical Prostatectomy: Are They Really Prostate Cancer Markers?, Maria Giulia Egidi, Giovanni Cochetti, Maria Rita Serva, Gabriella Guelfi, Danilo Zampini, Luca Mechelli, and Ettore -
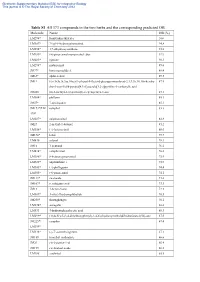
Table S1 All 373 Compounds in the Two Herbs and the Corresponding
Electronic Supplementary Material (ESI) for Integrative Biology This journal is © The Royal Society of Chemistry 2012 Table S1 All 373 compounds in the two herbs and the corresponding predicted OB Molecule Name OB (%) LM294* forsythidmethylester 100 LM387* 7'-epi-8-hydroxypinoresinol 94.8 LM394* 1,7-dihydroxyxanthone 93.8 LM289* (+)-pinoresinol monomethyl ether 92.9 LM285* egenine 90.3 LM298* matairesinol 89.6 JM77* loniceracetalide A 89.4 JM63* alpha-cedrol 89.3 JM1* (-)-(3r,8s,9r,9as,10as)-9-ethenyl-8-(beta-d-glucopyranosyloxy)-2,3,9,9a,10,10a-hexahy 87.5 dro-5-oxo-5h,8h-pyrano[4,3-d]oxazolo[3,2-a]pyridine-3-carboxylic acid JM200 (z)-3-methyl-2-(2-pentenyl)-2-cyclopenten-1-one 87.1 LM304* phillyrin 85.1 JM57* 7-epi-loganin 85.1 JM173*/LM camphol 83.3 398* LM287* epipinoresinol 82.8 JM27 2-methyl-1-butanol 81.2 LM356* (+)-lariciresinol 80.0 JM170* ledol 79.7 LM430 safynol 78.3 JM16 1-pentanol 76.2 LM414* campherenol 76.0 LM386* 8-hydroxypinoresinol 75.9 LM428* onjixanthone i 75.9 LM345* (+)-phillygenin 74.4 LM305* (+)-pinoresinol 74.1 JM113* sweroside 73.6 JM107* secologanic acid 73.3 JM13 1-hexen-3-one 72.3 LM309* 3-ethyl-7hydroxyphthalide 70.5 JM250* shuangkangsu 70.1 LM274* astragalin 68.6 LM331 4-hydroxyphenylacetic acid 68.3 LM299* (3r,4s,5r)-5-(3,4-dimethoxyphenyl)-3,4-bis(hydroxymethyl)dihydrofuran-2(3h)-one 67.5 JM225*/ camphor 67.4 LM399* LM338* (-)-7’-o-methylegenine 67.1 JM189 trimethyl isothiazole 66.6 JM29 cis-2-penten-1-ol 66.4 JM199 cis-linalool oxide 66.2 LM288 erythritol 65.8 Electronic Supplementary Material (ESI) for -

Systems Pharmacology Uncovers the Multiple Mechanisms of Xijiao Dihuang Decoction for the Treatment of Viral Hemorrhagic Fever
Hindawi Publishing Corporation Evidence-Based Complementary and Alternative Medicine Volume 2016, Article ID 9025036, 17 pages http://dx.doi.org/10.1155/2016/9025036 Research Article Systems Pharmacology Uncovers the Multiple Mechanisms of Xijiao Dihuang Decoction for the Treatment of Viral Hemorrhagic Fever Jianling Liu,1,2 Tianli Pei,1,2 Jiexin Mu,1,2 Chunli Zheng,2 Xuetong Chen,2 Chao Huang,2 Yingxue Fu,2 Zongsuo Liang,3 and Yonghua Wang2 1 CollegeofLifeScience,NorthwestUniversity,Xi’an,Shaanxi710069,China 2Center of Bioinformatics, College of Life Science, Northwest A&F University, Yangling, Shaanxi 712100, China 3College of Life Science, Zhejiang Sci-Tech University, Hangzhou, Zhejiang 310000, China Correspondence should be addressed to Yonghua Wang; yh [email protected] Received 21 December 2015; Revised 17 March 2016; Accepted 23 March 2016 Academic Editor: Hyunsu Bae Copyright © 2016 Jianling Liu et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Background. Viral hemorrhagic fevers (VHF) are a group of systemic diseases characterized by fever and bleeding, which have posed a formidable potential threat to public health with high morbidity and mortality. Traditional Chinese Medicine (TCM) formulas have been acknowledged with striking effects in treatment of hemorrhagic fever syndromes in China’s history. Nevertheless, their accurate mechanisms of action are still confusing. Objective. To systematically dissect the mechanisms of action of Chinese medicinal formula Xijiao Dihuang (XJDH) decoction as an effective treatment for VHF. Methods. In this study, a systems pharmacology method integrating absorption, distribution, metabolism, and excretion (ADME) screening, drug targeting, network, and pathway analysis was developed.