Bayesian Linear Regression Ahmed Ali, Alan N
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

DSA 5403 Bayesian Statistics: an Advancing Introduction 16 Units – Each Unit a Week’S Work
DSA 5403 Bayesian Statistics: An Advancing Introduction 16 units – each unit a week’s work. The following are the contents of the course divided into chapters of the book Doing Bayesian Data Analysis . by John Kruschke. The course is structured around the above book but will be embellished with more theoretical content as needed. The book can be obtained from the library : http://www.sciencedirect.com/science/book/9780124058880 Topics: This is divided into three parts From the book: “Part I The basics: models, probability, Bayes' rule and r: Introduction: credibility, models, and parameters; The R programming language; What is this stuff called probability?; Bayes' rule Part II All the fundamentals applied to inferring a binomial probability: Inferring a binomial probability via exact mathematical analysis; Markov chain Monte C arlo; J AG S ; Hierarchical models; Model comparison and hierarchical modeling; Null hypothesis significance testing; Bayesian approaches to testing a point ("Null") hypothesis; G oals, power, and sample size; S tan Part III The generalized linear model: Overview of the generalized linear model; Metric-predicted variable on one or two groups; Metric predicted variable with one metric predictor; Metric predicted variable with multiple metric predictors; Metric predicted variable with one nominal predictor; Metric predicted variable with multiple nominal predictors; Dichotomous predicted variable; Nominal predicted variable; Ordinal predicted variable; C ount predicted variable; Tools in the trunk” Part 1: The Basics: Models, Probability, Bayes’ Rule and R Unit 1 Chapter 1, 2 Credibility, Models, and Parameters, • Derivation of Bayes’ rule • Re-allocation of probability • The steps of Bayesian data analysis • Classical use of Bayes’ rule o Testing – false positives etc o Definitions of prior, likelihood, evidence and posterior Unit 2 Chapter 3 The R language • Get the software • Variables types • Loading data • Functions • Plots Unit 3 Chapter 4 Probability distributions. -

Generalized Linear Models and Generalized Additive Models
00:34 Friday 27th February, 2015 Copyright ©Cosma Rohilla Shalizi; do not distribute without permission updates at http://www.stat.cmu.edu/~cshalizi/ADAfaEPoV/ Chapter 13 Generalized Linear Models and Generalized Additive Models [[TODO: Merge GLM/GAM and Logistic Regression chap- 13.1 Generalized Linear Models and Iterative Least Squares ters]] [[ATTN: Keep as separate Logistic regression is a particular instance of a broader kind of model, called a gener- chapter, or merge with logis- alized linear model (GLM). You are familiar, of course, from your regression class tic?]] with the idea of transforming the response variable, what we’ve been calling Y , and then predicting the transformed variable from X . This was not what we did in logis- tic regression. Rather, we transformed the conditional expected value, and made that a linear function of X . This seems odd, because it is odd, but it turns out to be useful. Let’s be specific. Our usual focus in regression modeling has been the condi- tional expectation function, r (x)=E[Y X = x]. In plain linear regression, we try | to approximate r (x) by β0 + x β. In logistic regression, r (x)=E[Y X = x] = · | Pr(Y = 1 X = x), and it is a transformation of r (x) which is linear. The usual nota- tion says | ⌘(x)=β0 + x β (13.1) · r (x) ⌘(x)=log (13.2) 1 r (x) − = g(r (x)) (13.3) defining the logistic link function by g(m)=log m/(1 m). The function ⌘(x) is called the linear predictor. − Now, the first impulse for estimating this model would be to apply the transfor- mation g to the response. -

The Exponential Family 1 Definition
The Exponential Family David M. Blei Columbia University November 9, 2016 The exponential family is a class of densities (Brown, 1986). It encompasses many familiar forms of likelihoods, such as the Gaussian, Poisson, multinomial, and Bernoulli. It also encompasses their conjugate priors, such as the Gamma, Dirichlet, and beta. 1 Definition A probability density in the exponential family has this form p.x / h.x/ exp >t.x/ a./ ; (1) j D f g where is the natural parameter; t.x/ are sufficient statistics; h.x/ is the “base measure;” a./ is the log normalizer. Examples of exponential family distributions include Gaussian, gamma, Poisson, Bernoulli, multinomial, Markov models. Examples of distributions that are not in this family include student-t, mixtures, and hidden Markov models. (We are considering these families as distributions of data. The latent variables are implicitly marginalized out.) The statistic t.x/ is called sufficient because the probability as a function of only depends on x through t.x/. The exponential family has fundamental connections to the world of graphical models (Wainwright and Jordan, 2008). For our purposes, we’ll use exponential 1 families as components in directed graphical models, e.g., in the mixtures of Gaussians. The log normalizer ensures that the density integrates to 1, Z a./ log h.x/ exp >t.x/ d.x/ (2) D f g This is the negative logarithm of the normalizing constant. The function h.x/ can be a source of confusion. One way to interpret h.x/ is the (unnormalized) distribution of x when 0. It might involve statistics of x that D are not in t.x/, i.e., that do not vary with the natural parameter. -

Practical Statistics for Particle Physics Lecture 1 AEPS2018, Quy Nhon, Vietnam
Practical Statistics for Particle Physics Lecture 1 AEPS2018, Quy Nhon, Vietnam Roger Barlow The University of Huddersfield August 2018 Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 1 / 34 Lecture 1: The Basics 1 Probability What is it? Frequentist Probability Conditional Probability and Bayes' Theorem Bayesian Probability 2 Probability distributions and their properties Expectation Values Binomial, Poisson and Gaussian 3 Hypothesis testing Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 2 / 34 Question: What is Probability? Typical exam question Q1 Explain what is meant by the Probability PA of an event A [1] Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 3 / 34 Four possible answers PA is number obeying certain mathematical rules. PA is a property of A that determines how often A happens For N trials in which A occurs NA times, PA is the limit of NA=N for large N PA is my belief that A will happen, measurable by seeing what odds I will accept in a bet. Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 4 / 34 Mathematical Kolmogorov Axioms: For all A ⊂ S PA ≥ 0 PS = 1 P(A[B) = PA + PB if A \ B = ϕ and A; B ⊂ S From these simple axioms a complete and complicated structure can be − ≤ erected. E.g. show PA = 1 PA, and show PA 1.... But!!! This says nothing about what PA actually means. Kolmogorov had frequentist probability in mind, but these axioms apply to any definition. Roger Barlow ( Huddersfield) Statistics for Particle Physics August 2018 5 / 34 Classical or Real probability Evolved during the 18th-19th century Developed (Pascal, Laplace and others) to serve the gambling industry. -
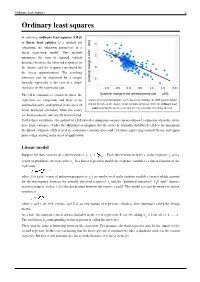
Ordinary Least Squares 1 Ordinary Least Squares
Ordinary least squares 1 Ordinary least squares In statistics, ordinary least squares (OLS) or linear least squares is a method for estimating the unknown parameters in a linear regression model. This method minimizes the sum of squared vertical distances between the observed responses in the dataset and the responses predicted by the linear approximation. The resulting estimator can be expressed by a simple formula, especially in the case of a single regressor on the right-hand side. The OLS estimator is consistent when the regressors are exogenous and there is no Okun's law in macroeconomics states that in an economy the GDP growth should multicollinearity, and optimal in the class of depend linearly on the changes in the unemployment rate. Here the ordinary least squares method is used to construct the regression line describing this law. linear unbiased estimators when the errors are homoscedastic and serially uncorrelated. Under these conditions, the method of OLS provides minimum-variance mean-unbiased estimation when the errors have finite variances. Under the additional assumption that the errors be normally distributed, OLS is the maximum likelihood estimator. OLS is used in economics (econometrics) and electrical engineering (control theory and signal processing), among many areas of application. Linear model Suppose the data consists of n observations { y , x } . Each observation includes a scalar response y and a i i i vector of predictors (or regressors) x . In a linear regression model the response variable is a linear function of the i regressors: where β is a p×1 vector of unknown parameters; ε 's are unobserved scalar random variables (errors) which account i for the discrepancy between the actually observed responses y and the "predicted outcomes" x′ β; and ′ denotes i i matrix transpose, so that x′ β is the dot product between the vectors x and β. -

Generalized and Weighted Least Squares Estimation
LINEAR REGRESSION ANALYSIS MODULE – VII Lecture – 25 Generalized and Weighted Least Squares Estimation Dr. Shalabh Department of Mathematics and Statistics Indian Institute of Technology Kanpur 2 The usual linear regression model assumes that all the random error components are identically and independently distributed with constant variance. When this assumption is violated, then ordinary least squares estimator of regression coefficient looses its property of minimum variance in the class of linear and unbiased estimators. The violation of such assumption can arise in anyone of the following situations: 1. The variance of random error components is not constant. 2. The random error components are not independent. 3. The random error components do not have constant variance as well as they are not independent. In such cases, the covariance matrix of random error components does not remain in the form of an identity matrix but can be considered as any positive definite matrix. Under such assumption, the OLSE does not remain efficient as in the case of identity covariance matrix. The generalized or weighted least squares method is used in such situations to estimate the parameters of the model. In this method, the deviation between the observed and expected values of yi is multiplied by a weight ω i where ω i is chosen to be inversely proportional to the variance of yi. n 2 For simple linear regression model, the weighted least squares function is S(,)ββ01=∑ ωii( yx −− β0 β 1i) . ββ The least squares normal equations are obtained by differentiating S (,) ββ 01 with respect to 01 and and equating them to zero as nn n ˆˆ β01∑∑∑ ωβi+= ωiixy ω ii ii=11= i= 1 n nn ˆˆ2 βω01∑iix+= βω ∑∑ii x ω iii xy. -

Applications of Computational Statistics with Multiple Regressions
International Journal of Computational and Applied Mathematics. ISSN 1819-4966 Volume 12, Number 3 (2017), pp. 923-934 © Research India Publications http://www.ripublication.com Applications of Computational Statistics with Multiple Regressions S MD Riyaz Ahmed Research Scholar, Department of Mathematics, Rayalaseema University, A.P, India Abstract Applications of Computational Statistics with Multiple Regression through the use of Excel, MATLAB and SPSS. It has been widely used for a long time in various fields, such as operations research and analysis, automatic control, probability statistics, statistics, digital signal processing, dynamic system mutilation, etc. The regress function command in MATLAB toolbox provides a helpful tool for multiple linear regression analysis and model checking. Multiple regression analysis is additionally extremely helpful in experimental scenario wherever the experimenter will management the predictor variables. Keywords - Multiple Linear Regression, Excel, MATLAB, SPSS, least square, Matrix notations 1. INTRODUCTION Multiple linear regressions are one of the foremost wide used of all applied mathematics strategies. Multiple regression analysis is additionally extremely helpful in experimental scenario wherever the experimenter will management the predictor variables [1]. A single variable quantity within the model would have provided an inadequate description since a number of key variables have an effect on the response variable in necessary and distinctive ways that [2]. It makes an attempt to model the link between two or more variables and a response variable by fitting a equation to determined information. Each value of the independent variable x is related to a value of the dependent variable y. The population regression line for p informative variables 924 S MD Riyaz Ahmed x1, x 2 , x 3 ...... -

1 Introduction to Bayesian Inference 2 Introduction to Gibbs Sampling
MCMC I: July 5, 2016 1 MCMC I 8th Summer Institute in Statistics and Modeling in Infectious Diseases Course Time Plan July 13-15, 2016 Instructors: Vladimir Minin, Kari Auranen, M. Elizabeth Halloran Course Description: This module is an introduction to Markov chain Monte Carlo methods with some simple applications in infectious disease studies. The course includes an introduction to Bayesian inference, Monte Carlo, MCMC, some background theory, and convergence diagnostics. Algorithms include Gibbs sampling and Metropolis-Hastings and combinations. Programming is in R. Familiarity with the R statistical package or other computing language is needed. Course schedule: The course is composed of 10 90-minute sessions, for a total of 15 hours of instruction. 1 Introduction to Bayesian Inference • Overview of the course. • Bayesian inference: Likelihood, prior, posterior, normalizing constant • Conjugate priors; Beta-binomial; Poisson-gamma; normal-normal • Posterior summaries, mean, mode, posterior intervals • Motivating examples: Chain binomial model (Reed-Frost), General Epidemic Model, SIS model. • Lab: – Goals: Warm-up with R for simple Bayesian computation – Example: Posterior distribution of transmission probability with a binomial sampling distribution using a conjugate beta prior distribution – Summarizing posterior inference (mean, median, posterior quantiles and intervals) – Varying the amount of prior information – Writing an R function 2 Introduction to Gibbs Sampling • Chain binomial model and data augmentation • Brief introduction -
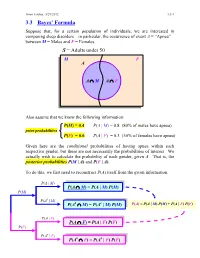
3.3 Bayes' Formula
Ismor Fischer, 5/29/2012 3.3-1 3.3 Bayes’ Formula Suppose that, for a certain population of individuals, we are interested in comparing sleep disorders – in particular, the occurrence of event A = “Apnea” – between M = Males and F = Females. S = Adults under 50 M F A A ∩ M A ∩ F Also assume that we know the following information: P(M) = 0.4 P(A | M) = 0.8 (80% of males have apnea) prior probabilities P(F) = 0.6 P(A | F) = 0.3 (30% of females have apnea) Given here are the conditional probabilities of having apnea within each respective gender, but these are not necessarily the probabilities of interest. We actually wish to calculate the probability of each gender, given A. That is, the posterior probabilities P(M | A) and P(F | A). To do this, we first need to reconstruct P(A) itself from the given information. P(A | M) P(A ∩ M) = P(A | M) P(M) P(M) P(Ac | M) c c P(A ∩ M) = P(A | M) P(M) P(A) = P(A | M) P(M) + P(A | F) P(F) P(A | F) P(A ∩ F) = P(A | F) P(F) P(F) P(Ac | F) c c P(A ∩ F) = P(A | F) P(F) Ismor Fischer, 5/29/2012 3.3-2 So, given A… P(M ∩ A) P(A | M) P(M) P(M | A) = P(A) = P(A | M) P(M) + P(A | F) P(F) (0.8)(0.4) 0.32 = (0.8)(0.4) + (0.3)(0.6) = 0.50 = 0.64 and posterior P(F ∩ A) P(A | F) P(F) P(F | A) = = probabilities P(A) P(A | M) P(M) + P(A | F) P(F) (0.3)(0.6) 0.18 = (0.8)(0.4) + (0.3)(0.6) = 0.50 = 0.36 S Thus, the additional information that a M F randomly selected individual has apnea (an A event with probability 50% – why?) increases the likelihood of being male from a prior probability of 40% to a posterior probability 0.32 0.18 of 64%, and likewise, decreases the likelihood of being female from a prior probability of 60% to a posterior probability of 36%. -

Ordinary Least Squares: the Univariate Case
Introduction The OLS method The linear causal model A simulation & applications Conclusion and exercises Ordinary Least Squares: the univariate case Clément de Chaisemartin Majeure Economie September 2011 Clément de Chaisemartin Ordinary Least Squares Introduction The OLS method The linear causal model A simulation & applications Conclusion and exercises 1 Introduction 2 The OLS method Objective and principles of OLS Deriving the OLS estimates Do OLS keep their promises ? 3 The linear causal model Assumptions Identification and estimation Limits 4 A simulation & applications OLS do not always yield good estimates... But things can be improved... Empirical applications 5 Conclusion and exercises Clément de Chaisemartin Ordinary Least Squares Introduction The OLS method The linear causal model A simulation & applications Conclusion and exercises Objectives Objective 1 : to make the best possible guess on a variable Y based on X . Find a function of X which yields good predictions for Y . Given cigarette prices, what will be cigarettes sales in September 2010 in France ? Objective 2 : to determine the causal mechanism by which X influences Y . Cetebus paribus type of analysis. Everything else being equal, how a change in X affects Y ? By how much one more year of education increases an individual’s wage ? By how much the hiring of 1 000 more policemen would decrease the crime rate in Paris ? The tool we use = a data set, in which we have the wages and number of years of education of N individuals. Clément de Chaisemartin Ordinary Least Squares Introduction The OLS method The linear causal model A simulation & applications Conclusion and exercises Objective and principles of OLS What we have and what we want For each individual in our data set we observe his wage and his number of years of education. -

Naïve Bayes Classifier
CS 60050 Machine Learning Naïve Bayes Classifier Some slides taken from course materials of Tan, Steinbach, Kumar Bayes Classifier ● A probabilistic framework for solving classification problems ● Approach for modeling probabilistic relationships between the attribute set and the class variable – May not be possible to certainly predict class label of a test record even if it has identical attributes to some training records – Reason: noisy data or presence of certain factors that are not included in the analysis Probability Basics ● P(A = a, C = c): joint probability that random variables A and C will take values a and c respectively ● P(A = a | C = c): conditional probability that A will take the value a, given that C has taken value c P(A,C) P(C | A) = P(A) P(A,C) P(A | C) = P(C) Bayes Theorem ● Bayes theorem: P(A | C)P(C) P(C | A) = P(A) ● P(C) known as the prior probability ● P(C | A) known as the posterior probability Example of Bayes Theorem ● Given: – A doctor knows that meningitis causes stiff neck 50% of the time – Prior probability of any patient having meningitis is 1/50,000 – Prior probability of any patient having stiff neck is 1/20 ● If a patient has stiff neck, what’s the probability he/she has meningitis? P(S | M )P(M ) 0.5×1/50000 P(M | S) = = = 0.0002 P(S) 1/ 20 Bayesian Classifiers ● Consider each attribute and class label as random variables ● Given a record with attributes (A1, A2,…,An) – Goal is to predict class C – Specifically, we want to find the value of C that maximizes P(C| A1, A2,…,An ) ● Can we estimate -
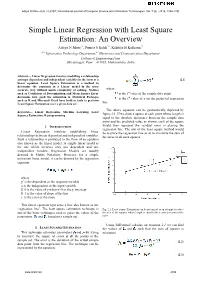
Simple Linear Regression with Least Square Estimation: an Overview
Aditya N More et al, / (IJCSIT) International Journal of Computer Science and Information Technologies, Vol. 7 (6) , 2016, 2394-2396 Simple Linear Regression with Least Square Estimation: An Overview Aditya N More#1, Puneet S Kohli*2, Kshitija H Kulkarni#3 #1-2Information Technology Department,#3 Electronics and Communication Department College of Engineering Pune Shivajinagar, Pune – 411005, Maharashtra, India Abstract— Linear Regression involves modelling a relationship amongst dependent and independent variables in the form of a (2.1) linear equation. Least Square Estimation is a method to determine the constants in a Linear model in the most accurate way without much complexity of solving. Metrics where such as Coefficient of Determination and Mean Square Error is the ith value of the sample data point determine how good the estimation is. Statistical Packages is the ith value of y on the predicted regression such as R and Microsoft Excel have built in tools to perform Least Square Estimation over a given data set. line The above equation can be geometrically depicted by Keywords— Linear Regression, Machine Learning, Least Squares Estimation, R programming figure 2.1. If we draw a square at each point whose length is equal to the absolute difference between the sample data point and the predicted value as shown, each of the square would then represent the residual error in placing the I. INTRODUCTION regression line. The aim of the least square method would Linear Regression involves establishing linear be to place the regression line so as to minimize the sum of relationships between dependent and independent variables.