Sequence and Phylogenetic Analysis of SSU Rrna Gene of Five Microsporidia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Sequence and Phylogenetic Analysis of SSU Rrna Gene of Five Microsporidia
Curr Microbiol (2010) 60:30–37 DOI 10.1007/s00284-009-9495-7 Sequence and Phylogenetic Analysis of SSU rRNA Gene of Five Microsporidia ShiNan Dong Æ ZhongYuan Shen Æ Li Xu Æ Feng Zhu Received: 21 May 2009 / Accepted: 24 August 2009 / Published online: 19 September 2009 Ó The Author(s) 2009. This article is published with open access at Springerlink.com Abstract The complete small subunit rRNA (SSU to elucidate phylogenetic relationships and taxonomic rRNA) gene sequences of five microsporidia including status of microsporidian species by analyzing information Nosema heliothidis, and four novel microsporidia isolated from SSU rRNA sequences of microsporidia. from Pieris rapae, Phyllobrotica armta, Hemerophila atrilineata, and Bombyx mori, respectively, were obtained by PCR amplification, cloning, and sequencing. Two Introduction phylogenetic trees based on SSU rRNA sequences had been constructed by using Neighbor-Joining of Phylip Microsporidia are obligate intracellular parasites that infect software and UPGMA of MEGA4.0 software. The taxo- a broad range of invertebrates and humans, they were nomic status of four novel microsporidia was determined recognized as a causative agent for many hosts including by analysis of phylogenetic relationship, length, G?C insects, fishes, rodents, and primates. In 1857, the first content, identity, and divergence of the SSU rRNA microsporidian species—N. bombycis was described by sequences. The results showed that the microsporidia iso- Nageli [1]. Until now, more than 1,200 microsporidia lated from Pieris rapae, Phyllobrotica armta, and He- species belonging to *150 genera have been reported merophila atrilineata have close phylogenetic relationship [2, 3]. with the Nosema, while another microsporidium isolated The adaptation of microsporidia to a parasitic lifestyle from Bombyx mori is closely related to the Endoreticulatus. -

Filling Gaps in the Microsporidian Tree: Rdna Phylogeny of Chytridiopsis Typographi (Microsporidia: Chytridiopsida)
Parasitology Research (2019) 118:169–180 https://doi.org/10.1007/s00436-018-6130-1 GENETICS, EVOLUTION, AND PHYLOGENY - ORIGINAL PAPER Filling gaps in the microsporidian tree: rDNA phylogeny of Chytridiopsis typographi (Microsporidia: Chytridiopsida) Daniele Corsaro1 & Claudia Wylezich2 & Danielle Venditti1 & Rolf Michel3 & Julia Walochnik4 & Rudolf Wegensteiner5 Received: 7 August 2018 /Accepted: 23 October 2018 /Published online: 12 November 2018 # Springer-Verlag GmbH Germany, part of Springer Nature 2018 Abstract Microsporidia are intracellular eukaryotic parasites of animals, characterized by unusual morphological and genetic features. They can be divided in three main groups, the classical microsporidians presenting all the features of the phylum and two putative primitive groups, the chytridiopsids and metchnikovellids. Microsporidia originated from microsporidia-like organisms belong- ing to a lineage of chytrid-like endoparasites basal or sister to the Fungi. Genetic and genomic data are available for all members, except chytridiopsids. Herein, we filled this gap by obtaining the rDNA sequence (SSU-ITS-partial LSU) of Chytridiopsis typographi (Chytridiopsida), a parasite of bark beetles. Our rDNA molecular phylogenies indicate that Chytridiopsis branches earlier than metchnikovellids, commonly thought ancestral, forming the more basal lineage of the Microsporidia. Furthermore, our structural analyses showed that only classical microsporidians present 16S-like SSU rRNA and 5.8S/LSU rRNA gene fusion, whereas the standard eukaryote rRNA gene structure, although slightly reduced, is still preserved in the primitive microsporidians, including 18S-like SSU rRNA with conserved core helices, and ITS2-like separating 5.8S from LSU. Overall, our results are consistent with the scenario of an evolution from microsporidia-like rozellids to microsporidians, however suggesting for metchnikovellids a derived position, probably related to marine transition and adaptation to hyperparasitism. -

Effects of Genotypic and Phenotypic Variation on Establishment Are Important for Conservation, Invasion, and Infection Biology
Effects of genotypic and phenotypic variation on establishment are important for conservation, invasion, and infection biology Anders Forsman1 Ecology and Evolution in Microbial Model Systems, Department of Biology and Environmental Science, Linnaeus University, SE-391 82 Kalmar, Sweden Edited by Ilkka Hanski, University of Helsinki, Helsinki, Finland, and approved December 2, 2013 (received for review September 20, 2013) There is abundant evidence that the probability of successful that more diverse groups harbor preadapted phenotypes, i.e., establishment in novel environments increases with number of a sampling effect (15, 16), niche complementarity resulting from individuals in founder groups and with number of repeated intro- reduced competition in groups where different phenotypes ex- ductions. Theory posits that the genotypic and phenotypic variation ploit different resources (17, 18), and facilitation, i.e., when the among individuals should also be important, but few studies have presence of one genotype or phenotype promotes the success of examined whether founder diversity influences establishment in- other phenotypes (19, 20). In addition, diverse populations of dependent of propagule pressure, nor whether the effect is model animals and plants may be less vulnerable to predators, diseases, or context dependent. I summarize the results of 18 experimental and pathogens (21, 22). Finally, genetic and phenotypic variation studies and report on a metaanalysis that provides strong evidence may promote population persistence because it buffers against that higher levels of genotypic and phenotypic diversity in founder selection in changing environments and enables adaptations to groups increase establishment success in plants and animals. The novel and changing conditions (10, 23–25). The above mecha- effect of diversity is stronger in experiments carried out under nisms are neither exhaustive nor mutually exclusive. -

Evolutionary and Functional Studies on Microsporidian ATP-Binding
Infection, Genetics and Evolution 68 (2019) 136–144 Contents lists available at ScienceDirect Infection, Genetics and Evolution journal homepage: www.elsevier.com/locate/meegid Evolutionary and functional studies on microsporidian ATP-binding T cassettes: Insights into the adaptation of microsporidia to obligated intracellular parasitism Qiang Hea, Charles R. Vossbrinckc, Qiong Yangd, Xian-Zhi Menga, Jian Luoa, Guo-Qing Pana, ⁎ ⁎ Ze-Yang Zhoua,b, , Tian Lia, a State Key Laboratory of Silkworm Genome Biology, Southwest University, Chongqing 400715, China b College of Life Science, Chongqing Normal University, Chongqing 400047, China c Department of Soil and Water, The Connecticut Agricultural Experiment Station, 123 Huntington Street, New Haven, CT 06511, USA d Sericulture and Agri-Food Research Institute, Guangdong Academy of Agricultural Sciences, Guangzhou, China ARTICLE INFO ABSTRACT Keywords: ATP-binding cassette (ABC) transporters comprise the largest family of transmembrane proteins and are found in Microsporidian all domains of life. The ABCs are involved in a variety of biological processes and as exporters play important ABC transporter roles in multidrug resistance. However, the ABC transporters have not been addressed in microsporidia, which Genome are a very large group of obligate intracellular parasites that can infect nearly all animals, including humans. Evolution Here, a total of 234 ABC transporters were identified from 18 microsporidian genomes and classified intofive Nosema bombycis subfamilies, including 74 ABCBs, 2 ABCCs, 18 ABCEs, 15 ABCFs, 102 ABCGs and 23 uncategorized members. Two subfamilies, ABCA and ABCD, are found in most organisms, but lost in microsporidia. Phylogenetic analysis indicated that microsporidian ABCB and ABCG subfamilies expanded by recent gene duplications, which re- sulted in the two largest subfamilies in microsporidia. -

Virtual 2-D Map of the Fungal Proteome
www.nature.com/scientificreports OPEN Virtual 2‑D map of the fungal proteome Tapan Kumar Mohanta1,6*, Awdhesh Kumar Mishra2,6, Adil Khan1, Abeer Hashem3,4, Elsayed Fathi Abd‑Allah5 & Ahmed Al‑Harrasi1* The molecular weight and isoelectric point (pI) of the proteins plays important role in the cell. Depending upon the shape, size, and charge, protein provides its functional role in diferent parts of the cell. Therefore, understanding to the knowledge of their molecular weight and charges is (pI) is very important. Therefore, we conducted a proteome‑wide analysis of protein sequences of 689 fungal species (7.15 million protein sequences) and construct a virtual 2‑D map of the fungal proteome. The analysis of the constructed map revealed the presence of a bimodal distribution of fungal proteomes. The molecular mass of individual fungal proteins ranged from 0.202 to 2546.166 kDa and the predicted isoelectric point (pI) ranged from 1.85 to 13.759 while average molecular weight of fungal proteome was 50.98 kDa. A non‑ribosomal peptide synthase (RFU80400.1) found in Trichoderma arundinaceum was identifed as the largest protein in the fungal kingdom. The collective fungal proteome is dominated by the presence of acidic rather than basic pI proteins and Leu is the most abundant amino acid while Cys is the least abundant amino acid. Aspergillus ustus encodes the highest percentage (76.62%) of acidic pI proteins while Nosema ceranae was found to encode the highest percentage (66.15%) of basic pI proteins. Selenocysteine and pyrrolysine amino acids were not found in any of the analysed fungal proteomes. -

Four QTL Underlie Resistance to a Microsporidian Parasite That May Drive Genome Evolution in Its 2 Daphnia Host
bioRxiv preprint doi: https://doi.org/10.1101/847194; this version posted November 20, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. 1 Four QTL underlie resistance to a microsporidian parasite that may drive genome evolution in its 2 Daphnia host 3 Devon Keller1, Devin Kirk1,2, Pepijn Luijckx1,3 4 1 Department of Ecology and Evolutionary Biology, University of Toronto, Toronto, Ontario, Canada, 5 M5S 3G5. 6 2 Current address: Department of Biology, Stanford University, Stanford, USA. 7 3 School of Natural Sciences, Zoology, Trinity College Dublin, Dublin 2, Ireland 8 Author contributions 9 DK, DK and PL designed the study and conducted experiments. PL conducted the analysis. PL wrote the 10 first draft of the manuscript, and all authors significantly contributed to revisions. 11 Abstract: 12 Despite its pivotal role in evolutionary and ecological processes the genetic architecture underlying host- 13 parasite interactions remains understudied. Here we use a quantitative trait loci approach to identify 14 regions in the Daphnia magna genome that provide resistance against its microsporidium parasite 15 Ordospora colligata. The probability that Daphnia became infected was affected by a single locus and an 16 interaction between two additional loci. A fourth locus influenced the number of spores that grew within 17 the host. Comparing our findings to previously published genetic work on Daphnia magna revealed that 18 two of these loci may be the same as detected for another microsporidium parasite, suggesting a general 19 immune response to this group of pathogens. -
![Ecology, Epidemiology, and Evolution of Parasitism in Daphnia [Internet]](https://docslib.b-cdn.net/cover/4327/ecology-epidemiology-and-evolution-of-parasitism-in-daphnia-internet-1394327.webp)
Ecology, Epidemiology, and Evolution of Parasitism in Daphnia [Internet]
Ecology, Epidemiology and Evolution of Parasitism in Daphnia Dieter Ebert Author’s address: Dieter Ebert Universität Basel Zoologisches Institut Evolutionsbiologie Vesalgasse 1 4051 Basel Switzerland Email: [email protected] For further information and updates please visit: http://evolution.unibas.ch/ Please cite as: Ebert D, 2005. Ecology, Epidemiology, and Evolution of Parasitism in Daphnia [Internet]. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. Available from: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Books Copyright: Please contact the author for permission to reproduce any or all of the content in this book. ISBN 1-932811-06-0 First published: 30 November 2005 (online version), 29 December 2005 (PDF version) The PDF version was produced by Thomas Zumbrunn, Universität Basel The cover illustration is part of a drawing from the monograph "Die günen Armpolypen, die geschwänzten und ungeschwänzten zackigen Wasserflöhe und eine besondere Art kleiner Wasser- aale" by Jacob Christian Schaffer, published in Regensburg, Germany in 1755. The picture shows the external and internal anatomy of Daphnia magna. Note that on the dorsal side the female carries numerous peritrich (Ciliata) epibionts. In the upper left corner a parthenogenetic egg is drawn. Ecology, Epidemiology, and Evolution of Parasitism in Daphnia Dieter Ebert Universität Basel Zoologisches Institut Evolutionsbiologie Vesalgasse 1 4051 Basel Switzerland http://evolution.unibas.ch/ Contents Acknowledgments xi 1 Introduction to the Ecology, Epidemiology, and Evolution of Parasitism in Daphnia 1 1.1 Foreword ............................................... 1 1.2 Setting the Stage ........................................... 1 1.3 Defining Parasites .......................................... 2 1.4 Host–Parasite Interactions ..................................... 2 1.5 Outline of This Book ....................................... -

Evolutionary Genomics of Metchnikovella Incurvata (Metchnikovellidae): an Early Branching Microsporidium
GBE Evolutionary Genomics of Metchnikovella incurvata (Metchnikovellidae): An Early Branching Microsporidium Luis Javier Galindo1,*, Guifre Torruella1, David Moreira1,Hele ` ne Timpano1, Gita Paskerova2,AlexeySmirnov2, Elena Nassonova2,3,andPurificacion Lopez-Garc ıa1,* 1Ecologie Systematique Evolution, CNRS, Universite Paris-Sud, AgroParisTech, Universite Paris-Saclay, Orsay, France 2Department of Invertebrate Zoology, Faculty of Biology, St Petersburg State University, Russia 3Laboratory of Cytology of Unicellular Organisms, Institute of Cytology Russian Academy of Sciences, St. Petersburg, Russia *Corresponding authors: E-mails: [email protected]; [email protected]. Accepted: September 12, 2018 Data deposition: This project has been deposited at DDBJ/ENA/GenBank under the BioProject number PRJNA477760, the SRA accession number SRP151413, and the Whole Genome Shotgun project QXFS00000000. Abstract Metchnikovellids are highly specialized hyperparasites, which infect and reproduce inside gregarines (Apicomplexa) inhab- iting marine invertebrates. Their phylogenetic affiliation was under constant discussion until recently, when analysis of the first near-complete metchnikovellid genome, that of Amphiamblys sp., placed it in a basal position with respect to most Microsporidia. Microsporidia are a highly diversified lineage of extremely reduced parasites related to Rozellida (Rozellosporidia ¼ Rozellomycota ¼ Cryptomycota) within the Holomycota clade of Opisthokonta. By sequencing DNA from a single-isolated infected gregarine cell we -

The Role of Microbiome and Genotype in Daphnia Magna Upon Parasite Re-Exposure
G C A T T A C G G C A T genes Article The Role of Microbiome and Genotype in Daphnia magna upon Parasite Re-Exposure Lore Bulteel 1,*, Shira Houwenhuyse 1 , Steven A. J. Declerck 2,3 and Ellen Decaestecker 1 1 Laboratory of Aquatic Biology, Department of Biology, University of Leuven-Campus Kulak, E. Sabbelaan 53, 8500 Kortrijk, Belgium; [email protected] (S.H.); [email protected] (E.D.) 2 Netherlands Institute of Ecology (NIOO-KNAW), Droevendaalsesteeg 10, 6700 AB Wageningen, The Netherlands; [email protected] 3 Laboratory of Aquatic Ecology, Evolution and Conservation, Department of Biology, KULeuven, 3000 Leuven, Belgium * Correspondence: [email protected]; Tel.: +32-4-9772-1255 Abstract: Recently, it has been shown that the community of gut microorganisms plays a crucial role in host performance with respect to parasite tolerance. Knowledge, however, is lacking on the role of the gut microbiome in mediating host tolerance after parasite re-exposure, especially considering multiple parasite infections. We here aimed to fill this knowledge gap by studying the role of the gut microbiome on tolerance in Daphnia magna upon multiple parasite species re-exposure. Additionally, we investigated the role of the host genotype in the interaction between the gut microbiome and the host phenotypic performance. A microbiome transplant experiment was performed in which three germ-free D. magna genotypes were exposed to a gut microbial inoculum and a parasite community treatment. The gut microbiome inocula were pre-exposed to the same parasite communities or a control treatment. Daphnia performance was monitored, and amplicon sequencing was performed to characterize the gut microbial community. -
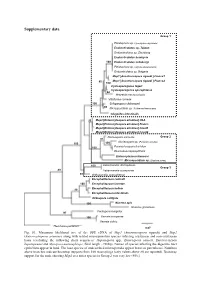
Supplementary Data
Supplementary data Group 1 Pleistophora sp. (Spodoptera depravata) Endoreticulatus sp. Taiwan Endoreticulatus sp. Zhenjiang Endoreticulatus bombycis 100 Endoreticulatus schubergi Pleistophora sp. (Agrotis exclamationis) Endoreticulatus sp. Bulgaria Msp1 [Anostracospora rigaudi ] France1 80 Msp1 [Anostracospora rigaudi ] France2 100 Cystosporogenes legeri Cystosporogenes operophterae 84 Mrazekia macrocyclopis Vittaforma corneae 100 Crispospora chironomi 80 Microsporidium sp. 3 (Artemia franciscana) Glugoides intestinalis 93 Msp2 [Enterocytospora artemiae] USA Msp2 [Enterocytospora artemiae] France Msp2 [Enterocytospora artemiae] Israel1 Msp2 [Enterocytospora artemiae] Israel2 Nucleospora salmonis Group 2 97 Nucleospora sp. (Parophrys vetulus) 100 100 100 Paranucleospora theridion Desmozoon lepeophtherii Enterocytozoon bieneusi Microsporidium sp. (Daphnia pulex) 100 Liebermannia dichroplusae Group 3 89 Liebermannia covasacrae 89 Orthosomella operophterae Encephalitozoon cuniculi 100 Encephalitozoon lacertae 99 Encephalitozoon hellem Encephalitozoon intestinalis Ordospora colligata 100 Nosema apis Nosema granulosis Facilispora margolisi 97 100 Vavraia oncoperae Vavraia culicis Thelohania parastaci 0.07 Fig. S1. Maximum likelihood tree of the SSU rDNA of Msp1 (Anostracospora rigaudi) and Msp2 (Enterocytospora artemiae) along with related microsporidian species infecting crustacean and non-crustacean hosts (excluding the following short sequences: Hepatospora spp., Enterospora canceri, Enterocytozoon hepatopenaei and Myospora metanephrops, -

Parasite-Mediated Selection in Experimental Metapopulations of Daphnia Magna Christoph R
Received 15 March 2004 Accepted 16 June 2004 Published online 22 September 2004 Parasite-mediated selection in experimental metapopulations of Daphnia magna Christoph R. Haag1,2Ãand Dieter Ebert1,2 1Universite´de Fribourg, De´partement de Biologie, Unite´d’Ecologie et d’Evolution, Chemin du Muse´e 10, CH-1700 Fribourg, Switzerland 2Tva¨rminne Zoological Station, SF-10900 Hanko, Finland In metapopulations, only a fraction of all local host populations may be infected with a given parasite species, and limited dispersal of parasites suggests that colonization of host populations by parasites may involve only a small number of parasite strains. Using hosts and parasites obtained from a natural metapopulation, we studied the evolutionary consequences of invasion by single strains of parasites in experimental populations of the cyclical parthenogen Daphnia magna. In two experiments, each spanning approximately one season, we monitored clone frequency changes in outdoor container populations consisting of 13 and 19 D. magna clones, respectively. The populations were either infected with single strains of the microsporidian parasites Octosporea bayeri or Ordospora colligata or left unparasitized. In both experiments, infection changed the representation of clones over time significantly, indicating parasite-mediated evolution in the experimental populations. Furthermore, the two parasite species changed clone frequencies differently, suggesting that the interaction between infection and competitive ability of the hosts was specific to the parasite species. Taken together, our results suggest that parasite strains that invade local host populations can lead to evolutionary changes in the genetic composition of the host population and that this change is parasite- species specific. Keywords: clonal competition; coevolution; genetic diversity; Microsporidia; resistance 1. -

Transposable Element Abundance Correlates with Mode Of
Albuquerque et al. Mobile DNA (2020) 11:19 https://doi.org/10.1186/s13100-020-00218-8 SHORT REPORT Open Access Transposable element abundance correlates with mode of transmission in microsporidian parasites Nathalia Rammé Medeiros de Albuquerque1, Dieter Ebert2 and Karen Luisa Haag1* Abstract The extreme genome reduction and physiological simplicity of some microsporidia has been attributed to their intracellular, obligate parasitic lifestyle. Although not all microsporidian genomes are small (size range from about 2 to 50 MB), it is suggested that the size of their genomes has been streamlined by natural selection. We explore the hypothesis that vertical transmission in microsporidia produces population bottlenecks, and thus reduces the effectiveness of natural selection. Here we compare the transposable element (TE) content of 47 microsporidian genomes, and show that genome size is positively correlated with the amount of TEs, and that species that experience vertical transmission have larger genomes with higher proportion of TEs. Our findings are consistent with earlier studies inferring that nonadaptive processes play an important role in microsporidian evolution. Background gene-sparse genomes is populated by repetitive se- Microsporidia, a phylum of fast evolving intracellular quences, most of which have been identified as transpos- parasites related to fungi [1–3], are interesting models able elements [11]. It is suggested that the majority of for studying the mechanisms shaping eukaryotic genome transposable element (TE) insertions cause mildly dele- evolution. Their genomes are small and highly diverse. terious effects in eukaryotes [12, 13]. Therefore, their Two contrasting genome architectures are found in spread and accumulation depend on the cellular mecha- microsporidians.