Stable Isotope Analysis of Ozark Hellbender (Cryptobranchus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Fishing on the Eleven Point River
FISHING ON THE ELEVEN POINT RIVER Fishing the Eleven Point National Scenic River is a very popular recreation activity on the Mark Twain National Forest. The river sees a variety of users and is shared by canoes and boats, swimmers, trappers, and anglers. Please use caution and courtesy when encountering another user. Be aware that 25 horsepower is the maximum boat motor size allowed on the Eleven Point River from Thomasville to "the Narrows" at Missouri State Highway 142. Several sections of the river are surrounded by private land. Before walking on the bank, ask the landowners for permission. Many anglers today enjoy the sport of the catch and fight, but release the fish un-harmed. Others enjoy the taste of freshly caught fish. Whatever your age, skill level or desire, you should be aware of fishing rules and regulations, and a little natural history of your game. The Varied Waters The Eleven Point River, because of its variety of water sources, offers fishing for both cold and warm-water fish. Those fishing the waters of the Eleven Point tend to divide the river into three distinctive areas. Different fish live in different parts of the river depending upon the water temperature and available habitat. The upper river, from Thomasville to the Greer Spring Branch, is good for smallmouth bass, longear sunfish, bluegill, goggle-eye (rock bass), suckers, and a few largemouth bass. This area of the river is warmer and its flow decreases during the summer. The river and fish communities change where Greer Spring Branch enters the river. -
Activity Guide 75+ Things to See and Do
Activity Guide 75+ Things to See and Do in and around Pocahontas and Randolph County Arkansas ! USA Experience The Lesmeister, a well- appointed retreat offering a restful escape from your busy life, or an active time spent seeing and doing in a classic American hill town. The choice is yours! • For ambiance, each living area has either a wood-burning stove or gas fireplace. • Bedrooms feature king size beds with high-thread count sheets • Enjoy private bathrooms with heated floors, ultra-clean Sanijet spa tubs, a shower, flat screen TV, towel warmer, toilet with heated seat/Bidet with warmed water, and a skylight to let the natural light come in. Make Secure Online Reservations 24/7 at ArkansasGuestHouse.com 208 N Marr Street, Pocahontas, Arkansas 501-291-1233 The perfect base for a visit to historic Pocahontas! Activity Guide Copyright © 2012-17 by ARSoft LLC. All rights reserved. TABLE OF CONTENTS Things to See & Do in Downtown Pocahontas Historic District Walking Tour. 7 NoMa Art and Entertainment District. 7 Downtown Playhouse.. 7 Bella Piazza Italian Restaurant. 8 Carroll’s Variety Store and Flea Market.. 8 1872 Randolph County Courthouse.. 9 Capture of Confederate General Jeff Thompson. 9 Arkansas’ Only Quilt Trail. 10 Studio B Salon and Day Spa. 11 Black River Beads, Pottery, and Glass Blowing. 11 Randolph County Heritage Museum. 11 The Treasure Trunk Eclectic Shopping. 12 Black River Overlook Park, Lover’s Lock Lane, and The Pocahontas Civil War River Walk Memorial. 12 Futrell’s Old Time Hardware. 14 R. J. Reynolds-Pearcy Art Gallery. 14 Arkansas’ Oldest Barber Shop. -

Fishes of Randolph County, Arkansas Steve M
Journal of the Arkansas Academy of Science Volume 31 Article 8 1977 Fishes of Randolph County, Arkansas Steve M. Bounds Arkansas State University John K. Beadles Arkansas State University Billy M. Johnson Arkansas State University Follow this and additional works at: http://scholarworks.uark.edu/jaas Part of the Aquaculture and Fisheries Commons, and the Terrestrial and Aquatic Ecology Commons Recommended Citation Bounds, Steve M.; Beadles, John K.; and Johnson, Billy M. (1977) "Fishes of Randolph County, Arkansas," Journal of the Arkansas Academy of Science: Vol. 31 , Article 8. Available at: http://scholarworks.uark.edu/jaas/vol31/iss1/8 This article is available for use under the Creative Commons license: Attribution-NoDerivatives 4.0 International (CC BY-ND 4.0). Users are able to read, download, copy, print, distribute, search, link to the full texts of these articles, or use them for any other lawful purpose, without asking prior permission from the publisher or the author. This Article is brought to you for free and open access by ScholarWorks@UARK. It has been accepted for inclusion in Journal of the Arkansas Academy of Science by an authorized editor of ScholarWorks@UARK. For more information, please contact [email protected], [email protected]. ! Journal of the Arkansas Academy of Science, Vol. 31 [1977], Art. 8 Fishes ofRandolph County, Arkansas STEVE M. BOUNDS,' JOHN K.BEADLESand BILLYM.JOHNSON Divisionof Biological Sciences, Arkansas State University I State University, Arkansas 72467 ! ABSTRACT Asurvey of the fishes of Randolph County in northcentral Arkansas was made between June 1973 and March 1977. Field collections, literature records, and museum specimens re- n vealed the ichthyofauna of Randolph County to be composed of 128 species distributed among 24 families. -

Missouri Conservationist July 2021
VOLUME 82, ISSUE 7, JULY 2021 MISSOURI SERVING NATURE & YOU CONSERVATIONIST VIEWING W ILDLIFE HUNTING CAMPING LOUNGING RUNNING & HIKING BIKING ARCHERY & SHOO TING STARGAZING FISHING SWIMMING mdc.mo.gov/NeverLoseTouch For ways to reconnect with nature, visit visit nature, with reconnect to ways For . Never Lose Touch Lose Never . and again, It’s time to make that connection connection that make to time It’s nature and exploring Missouri. exploring and nature to give back while being out in in out being while back give to nature. There are so many ways ways many so are There nature. VIEWING W ILDLIFE HUNTING CAMPING LOUNGING RUNNING & HIKING to helping people connect with with connect people helping to up streams and planting trees trees planting and streams up efforts in Missouri, from cleaning cleaning from Missouri, in efforts huge impact on conservation conservation on impact huge it thriving. Volunteers make a a make Volunteers thriving. it needs to be cared for to keep keep to for cared be to needs Nature is amazing. It also also It amazing. is Nature with nature? with time you connected connected you time When was the last last the was When BIKING ARCHERY & SHOO TING STARGAZING FISHING SWIMMING MISSOURI CONSERVATIONIST JULY 2021 Contents VOLUME 82, ISSUE 7 10 ON THE COVER Bushwhacker Lake Conservation Area : DAVID STONNER 16–35mm lens, f/11 1/15 sec, ISO 100 GOVERNOR Michael L. Parson THE CONSERVATION COMMISSION CHAIR Don C. Bedell VICE CHAIR Wm. L. (Barry) Orscheln SECRETARY Mark L. McHenry MEMBER Steven D. Harrison DIRECTOR Sara -

Fishes of the Eleven Point River Within Arkansas Michael B
Journal of the Arkansas Academy of Science Volume 31 Article 19 1977 Fishes of the Eleven Point River Within Arkansas Michael B. Johnson Arkansas State University John K. Beadles Arkansas State University Follow this and additional works at: http://scholarworks.uark.edu/jaas Part of the Aquaculture and Fisheries Commons, and the Terrestrial and Aquatic Ecology Commons Recommended Citation Johnson, Michael B. and Beadles, John K. (1977) "Fishes of the Eleven Point River Within Arkansas," Journal of the Arkansas Academy of Science: Vol. 31 , Article 19. Available at: http://scholarworks.uark.edu/jaas/vol31/iss1/19 This article is available for use under the Creative Commons license: Attribution-NoDerivatives 4.0 International (CC BY-ND 4.0). Users are able to read, download, copy, print, distribute, search, link to the full texts of these articles, or use them for any other lawful purpose, without asking prior permission from the publisher or the author. This Article is brought to you for free and open access by ScholarWorks@UARK. It has been accepted for inclusion in Journal of the Arkansas Academy of Science by an authorized editor of ScholarWorks@UARK. For more information, please contact [email protected], [email protected]. Journal of the Arkansas Academy of Science, Vol. 31 [1977], Art. 19 Fishes of the Eleven Point River Within Arkansas B.MICHAELJOHNSON and JOHN K.BEADLES Division of Biological Sciences Arkansas State University State University, Arkansas 72467 ABSTRACT A survey of the fishes of the Eleven Point River and its tributaries was made between 31 January 1976 and 13 February 1977. -

Water Resources of Randolph and Lawrence Counties, Arkansas
Water Resources of Randolph and Lawrence Counties, Arkansas GEOLOGICAL SURVEY WATER-SUPPLY PAPER 1879-B Prepared in cooperation with the Arkansas Geological Commission Water Resources of Randolph and Lawrence Counties, Arkansas By A. G. LAMONDS, MARION S. MINES, and RAYMOND 0. PLEBUCH CONTRIBUTIONS TO THE HYDROLOGY OF THE UNITED STATES GEOLOGICAL SURVEY WATER-SUPPLY PAPER 1879-B Prepared in cooperation with the Arkansas Geological Commission UNITED STATES GOVERNMENT PRINTING OFFICE, WASHINGTON : 1969 UNITED STATES DEPARTMENT OF THE INTERIOR WALTER J. HICKEL, Secretary GEOLOGICAL SURVEY William T. Pecora, Director For sale by the Superintendent of Documents, U.S. Government Pr'nting Office Washington, D.C. 20402 - Price 30 cents (paper cover) CONTENTS Page Abstract_ ----_-__--_--_--_-_-_--_-____-___----________-_____.-_- Bl Introduction._____________________________________________________ 2 Purpose.__--_____--_-____-__--________________________-___-_- 2 Acknowledgments. _ ___________________________________________ 2 The area.____________________________________________________ ? Surface-water resources.._-----____-_-_-_-___-_-_-_______________--_ ? Availability. __________________________________________________ fi Low-flow frequency._______________________________________ 5 Flow duration___________________________________________ 10 Floods.__________________________________________________ 12 Quality. ________------______--_-____._---____._-__-____-_-_- 14 Chemical and physical quality.-_____-_-___-___-_-_-_____-__ 14 Bacterial quality-_________________________________________ -

Species Status Assessment for Spring River Crayfish
Species Status Assessment Report for the Spring River Crayfish ( Faxonius roberti) Spring River Crayfish; Photo: Christopher Taylor, Illinois Natural History Survey U.S. Fish and Wildlife Service December 7, 2018 Acknowledgements This report was prepared by Trisha Crabill (Missouri Ecological Services Field Office), Laura Ragan (Midwest Regional Office), and Jonathan JaKa (U.S. Fish & Wildlife Service Headquarters) with assistance from Alyssa Bangs (Arkansas Ecological Services Field Office) and the following individuals from the Missouri Ecological Services Field Office: Scott Hamilton, Joshua Hundley, Ashton Jones, and Kaitlyn Kelly. We greatly appreciate the species experts who provided data and extensive input on various aspects of the SSA analysis, including a technical review of the draft report: Robert DiStefano (Missouri Department of Conservation), Dr. Daniel Magoulick (University of Arkansas), Dr. Christopher Taylor (I llinois Natural History Survey) , Brian Wagner (Arkansas Game and Fish Commission), and Dr. Jacob Westhoff (Missouri Department of Conservation). We also thank Dr. James Fetzner (Carnegie Museum of Natural History) for reviewing the draft report and providing information on the Faxonius wagneri a nd F. roberti s pecies delineations. Lastly, we thank Dr. Zachary Loughman (West Liberty University) and Christopher Rice (Missouri Department of Conservation) for providing a technical review of the draft report. Suggested citation: U.S. Fish and Wildlife Service. 2018. Species status assessment report for the Spring River Crayfish (F axonius roberti) . Version 1.0, December 2018. Midwest Region, Bloomington, Minnesota. 64 pp. 2 Executive Summary This report summarizes results of a species status assessment (SSA) conducted for the Spring River Crayfish (F axonius roberti) to assess its viability. -

Reports & Statistics
REPORTS & STATISTICS COLOSSIANS 1:1523 2019 MISSOURI BAPTIST CONVENTION ANNUAL MEETING 1 2 ANNUAL REPORT of the MISSOURI BAPTIST CONVENTION 185th ANNUAL MEETING Branson Convention Center October 28-29, 2019 v E X E C U T I V E D I R E C T O R Dr. John L. Yeats O F F I C E R S Jeremy Muniz, President Jon Nelson, First Vice President Jeff Anderson, Second Vice President Chad Hodges, Recording Secretary A S S I S TA N T T O T H E R E C O R D I N G S E C R E TA RY Carla Stegeman For more information, contact: Missouri Baptist Convention, 400 E High St, Jefferson City, Missouri 65101‑3215 Phone: 573‑636‑0400 Toll‑free: 800‑736‑6227 FAX: 573‑659‑7436 Copyright © 2020 Missouri Baptist Convention. All rights reserved. This book or any portion thereof may not be reproduced or used in any manner whatsoever without the express written permission of the publisher. The Annual is not available for use in developing mailing lists. A PDF file of the Annual is available for free download at: mobaptist.org/executive‑office/annual‑reports‑statistics 3 4 Table of Contents Section I Records of the Annual Meeting Proceedings Provisional Program Recommendations Resolutions Section II Audits Executive Board of the Missouri Baptist Convention Hannibal-LaGrange University Missouri Baptist Children’s Home Missouri Baptist Foundation Southwest Baptist University Section III Historical Information The Record (ACP Summaries) Missouri Baptist Associations and Regions (map) Resident Membership by Region and Association Statistics: Regions and Churches Summary of Statistics Record of Annual Meetings Section IV Denominational Directories Executive Board Officers and Members Boards of Benevolent Institutions Boards of Educational Institutions Boards of Agencies and Commissions 5 6 Section I Records of the Annual Meeting Proceedings Provisional Program Recommendations Resolutions The Proceedings and Provisional Program are printed as required by the Constitution of the Missouri Baptist Convention, Article VI — Annual Meeting, #5. -
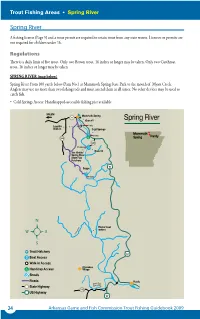
Spring River
Spring River SALEM Mammoth Spring 9 Dam #1 Mammoth Lassiter 1 mile Spring Hardy Trout FishingWalk-in Areas • Spring River Cold Springs Access Spring River 2 miles 289 A fishing license (Page 5) and a trout permit are required to retain trout 342from any state waters. Licenses or permits are not required for children under 16. 3.5 miles Regulations Dam #3 Jim Hinkle/ There is a daily limit of five trout. Only twoSpring Brown trout, River 16 inches or longer may be taken. Only two Cutthroat trout, 16 inches or longer may be taken. State Fish Spring River SALEM MammothHatchery Spring Spring river (map below) 9 Dam #1 Spring River From 100 yards below Dam No.1 at Mammoth Spring State Park Mammothto the mouth of63 Myatt Creek. Anglers may use Lassiterno more than two fishing rods1 mileand must attendBayou them at all times.Spring No otherHardy devices may be used to Walk-in Cold Springs catch fish. Access • Cold Springs Access: Handicapped-accessible fishing2 miles pier available. 289 6 miles 342 Spring River SpringSALEM River SALEM 3.5 miles Mammoth Spring Mammoth Spring 9 9 DamDam #1 #3 Spring River SALEM Jim Hinkle/ Dam #1 Spring River Mammoth Spring Mammoth SALEM LassiterSpring9 River 1 mile Hardy Mammoth Spring Mammoth Spring 9 1 mile Walk-inHardyState Fish Dam #1Cold Springs Lassiter Spring AccessHatchery Walk-in Dam #1 Mammoth Cold Springs 1 mile 2 miles Hardy Access Mammoth Lassiter 289 Spring 1 mile Walk-inHardy Cold Springs63 Lassiter 2 miles Spring Bayou 342 Walk-in Cold Springs Access 289 3.5 miles Access 2 miles Dam #3 2 miles -

Missouri Conservationist July 2019
VOLUME 80, ISSUE 2, FEBRUARY 2019 MISSOURI SERVING NATURE & YOU CONSERVATIONIST NATUREis Healthy Feeling tired? Spending Getting away from Taking a nature Exposure to nature Spending just 20 time in nature, busy schedules walk may increase contributes to minutes outside conservation allows people to attention spans physical well- can give your brain areas, woods, connect with and creative being, reducing an energy boost backyards, and nature and problem-solving blood pressure, comparable to a urban parks themselves in a skills by as much heart rate, muscle cup of coffee. may ease way that brings as 50 percent. tension, and the stress levels. calm and a sense production of of well-being. stress hormones. Get healthy in nature this year. Visit mdc.mo.gov/places-go or download the free MO Outdoors app for ideas on where to go near you. Download for Android MISSOURI CONSERVATIONIST FEBRUARY 2019 Contents VOLUME 80, ISSUE 2 10 ON THE COVER Gray squirrel : NOPPADOL PAOTHONG 800mm lens +1.4 teleconverter, f/11, 1/500 sec, ISO 800 GOVERNOR Michael L. Parson THE CONSERVATION COMMISSION CHAIR Marilynn J. Bradford VICE CHAIR David W. Murphy SECRETARY Nicole E. Wood MEMBER Don C. Bedell DIRECTOR Sara Parker Pauley DEPUTY DIRECTORS Mike Hubbard, Aaron Jeffries, Jennifer Battson Warren MAGAZINE STAFF EDITOR Angie Daly Morfeld ASSOCIATE EDITOR Bonnie Chasteen STAFF WRITERS Larry Archer, Heather Feeler, Kristie Hilgedick, Joe Jerek CREATIVE DIRECTOR Stephanie Thurber ART DIRECTOR Cliff White DESIGNERS Les Fortenberry, Marci Porter FEATURES PHOTOGRAPHERS Noppadol Paothong, David Stonner 10 CIRCULATION MANAGER Schoolcraft: 20 Laura Scheuler A Journey Through mdc.mo.gov/conmag Southern Missouri DEPARTMENTS Retracing the geographer’s historic trek and what it means today. -

Recent Collections of Fishes from the Spring River Drainage in Northeast Arkansas Steve C
Journal of the Arkansas Academy of Science Volume 41 Article 25 1987 Recent Collections of Fishes from the Spring River Drainage in Northeast Arkansas Steve C. Baker Arkansas Game and Fish Commission Michael L. Armstrong Arkansas Game and Fish Commission Follow this and additional works at: http://scholarworks.uark.edu/jaas Part of the Aquaculture and Fisheries Commons Recommended Citation Baker, Steve C. and Armstrong, Michael L. (1987) "Recent Collections of Fishes from the Spring River Drainage in Northeast Arkansas," Journal of the Arkansas Academy of Science: Vol. 41 , Article 25. Available at: http://scholarworks.uark.edu/jaas/vol41/iss1/25 This article is available for use under the Creative Commons license: Attribution-NoDerivatives 4.0 International (CC BY-ND 4.0). Users are able to read, download, copy, print, distribute, search, link to the full texts of these articles, or use them for any other lawful purpose, without asking prior permission from the publisher or the author. This General Note is brought to you for free and open access by ScholarWorks@UARK. It has been accepted for inclusion in Journal of the Arkansas Academy of Science by an authorized editor of ScholarWorks@UARK. For more information, please contact [email protected], [email protected]. Journal of the Arkansas Academy of Science, Vol. 41 [1987], Art. 25 GENERAL NOTES RECENT COLLECTIONS OF FISHES FROM THE SPRING RIVER DRAINAGE INNORTHEAST ARKANSAS Several ichthyofaunal surveys have been conducted in the Spring River of Arkansas. Meek (1894) initiallycollected fish insome tributaries of the Upper Spring River. Buchanan (1973) sampled throughout the drainage in developing his Key to the Fishes of Arkansas. -

Rivers: Revised November 2008; Caves: Revised July 2007; Trails: Revised March 2010; High Adventure: Revised September 2007
Everything you need to know about more than 90 of the best camps in the region Including more than 60 High Adventure opportunities Images courtesy of: http://signal.baldwincity.com/news/2011/oct/20/local-boy-scouts-troop-remained-busy-during-summer/ http://i4.ytimg.com/vi/obn8RVY_szM/mgdefault.jpg http://www/sccovington.com/philmont/trek_info/equipment/tents.htm This is a publication of Tamegonit Lodge, the Order of the Arrow lodge affiliated with the Heart of America Council, BSA. Updated: December 2012 Additional copies of this publication are available through the Program Services Department at the Heart of America Council Scout Service Center 10210 Holmes Road Kansas City, Missouri 64131 Phone: (816) 942-9333 Toll Free: (800) 776-1110 Fax: (816) 942-8086 Online: www.hoac-bsa.org Camps: Revised December 2012; Rivers: Revised November 2008; Caves: Revised July 2007; Trails: Revised March 2010; High Adventure: Revised September 2007 HOAC – Order of the Arrow – ON THE LOOSE RIVERS – Page 1 Welcome to the adventures which the scenic rivers in southern Missouri offer. In the next pages many rivers are described, both in general and by specific sections. You will also find within this section a compilation of many public outfitters, which regularly provide all necessary equipment for a canoe trip. The river sections that are profiled are probably not the exact sections of river that you will float if you rent your canoes and equipment form an outfitter. Each outfitter has “normal” floats that you may choose from, and few, if any, of these floats will match with the sections profiled in On The Loose, or with the sections any other outfitter would float! After spending some time with the River Section of On The Loose, I feel that you will agree there are many more rivers, which can be floated than the ones commonly mentioned (i.e.