Potential Biomarkers As an Indicator of Vertical Transmission of Johne's
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Giardia Intestinalis, Also Known As G
For Vets General Information Giardiosis is a disease caused by the protozoan parasite Giardia intestinalis, also known as G. duodenalis or G. lamblia. It is the most commonly identified pathogen in outbreaks of waterborne human disease in the USA. Clinical disease typically manifests as diarrhea. By far the most common source of infection for people is surface water contaminated by human fecal material. Within G. intestinalis there are a number of genotypes which are grouped into “assemblages” A to F. Assemblages A and B are capable of infecting several animal species, as well as humans. Other assemblages occur only in animals. Giardia, including zoonotic and species-specific assemblages, occurs frequently in dogs, and less commonly in cats, but the parasite can infect many species including beavers, livestock, ferrets, guinea pigs, gerbils, rats and chinchillas. The risk of zoonotic transmission of Giardia from pets remains controversial and unquantified, but the potential certainly exists. It is therefore important to be aware of the potential risk and encourage pet owners and people who handle animals to take steps to help prevent the spread of Giardia. Prevalence and Risk Factors Humans Giardiosis is endemic worldwide. In industrialized countries the prevalence of Giardia in people is estimated to be between 1-7%, but it may be as high as 50% in developing countries. All age groups are equally affected during epidemics, but both subclinical infection and clinical disease are more common in children in endemic areas. Outbreaks occur regularly in childcare facilities. Immunocompromised individuals are also more commonly affected than members of the general population. -

Asymptomatic Subclinical Cases of Coronavirus Disease 2019 Without Viral Transmission in Three Independent Families
Infection and Drug Resistance Dovepress open access to scientific and medical research Open Access Full Text Article SHORT REPORT Asymptomatic Subclinical Cases of Coronavirus Disease 2019 without Viral Transmission in Three Independent Families This article was published in the following Dove Press journal: Infection and Drug Resistance Xian Zhang1,* Purpose: There is increasing evidence indicating that considerable fractions of cases of Liting Chen2,* SARS-CoV-2 infection are asymptomatic. We traced three asymptomatic clusters to inves Jia Wei2 tigate the infectivity of subclinical cases of coronavirus disease 2019 (COVID-19). Jianfeng Zhou2 Patients and Methods: Three medical staff who were asymptomatic were diagnosed with Yang Cao2 coronavirus disease 2019 by serological tests. Their close contacts were systematically evaluated based on COVID-19-related symptoms, nucleic acid tests, serological tests, and Gaoxiang Wang2 chest computed tomography (CT) as needed to determine if they were infected by SARS- 1Department of Ophthalmology, Tongji CoV-2. Hospital, Tongji Medical College, Huazhong University of Science and Results: None of the staff’s close contacts, including 10 family members, were infected by Technology, Wuhan, Hubei 430030, the indexes, even though no protective measures were taken. 2 People’s Republic of China; Department Conclusion: The infectivity of asymptomatic subclinical infection patients of coronavirus of Hematology, Tongji Hospital, Tongji Medical College, Huazhong University of disease 2019 seems to be low. Science and Technology, Wuhan, Hubei Keywords: SARS-CoV-2, COVID-19, asymptomatic, close contact, infectivity 430030, People’s Republic of China *These authors contributed equally to this work Introduction Toward the end of 2019, there was an outbreak of severe acute respiratory syn drome coronavirus 2 (SARS-CoV-2). -

Subclinical Avian Influenza A(H5N1) Virus Infection in Human, Vietnam
DISPATCHES leading to hospital admission. Despite intensive care and Subclinical Avian treatment with oseltamivir and antibiotics, the disease pro- gressed, and he died 2 days later. Influenza A(H5N1) A throat swab taken from the index case-patient on day 3 of illness was tested by reverse transcription PCR, and Virus Infection in results were positive for influenza A(H5N1) virus. Hemag- Human, Vietnam glutination inhibition (HI) and microneutralization (MN) tests for H5N1-specific antibodies were negative in sam- Mai Quynh Le, Peter Horby, Annette Fox, ples taken during the acute phase of illness (online Techni- Hien Tran Nguyen, Hang Khanh Le Nguyen, cal Appendix, wwwnc.cdc.gov/EID/article/19/10/13-0730- Phuong Mai Vu Hoang, Khanh Cong Nguyen, Techapp1.pdf). Menno D. de Jong, Rienk E. Jeeninga, On day 5 of illness of the index case-patient, a contact H. Rogier van Doorn, Jeremy Farrar, investigation was initiated. Throat swab specimens were and Heiman F.L. Wertheim collected from 4 household members and 1 close contact of the index case-patient: his spouse (age 47 years), daughter Laboratory-confirmed cases of subclinical infection (age 18 years), daughter-in-law (age 25 years), and grand- with avian influenza A(H5N1) virus in humans are rare, and son (age 1 year) and an unrelated man (age 43 years). None the true number of these cases is unknown. We describe of the contacts had signs or symptoms. Infection control the identification of a laboratory-confirmed subclinical case measures were initiated, and all household members were in a woman during an influenza A(H5N1) contact investiga- tion in northern Vietnam. -

Management of Water-Related Microbial Diseases
Management of Water-related Microbial Diseases First Edition 2003 The Department of Water U Affairs and Forestry Tl I75/DJ Water Research Commission REPORTS IN THIS SERIES This guide forms part of a series which is aimed at water supply agencies, water resource managers, workers in health-related fields, as well as communities throughout South Africa. The guide is intended to provide awareness-building information to keep water supplies clean of microbial contamination and thus reduce the incidence of water-related diseases. The publication of this report emanates from a WRC consultancy no 431: Guide on water-related microbiological diseases. The following documents form part of this series of Guides on the Management of Water-related Microbial Diseases: Vol: 1 What is the problem? - Disease Characteristics. Vol: 2 What causes the problem;1 - A What to Do for Water Suppliers following Diarrhoea Incidents. ' Vol: 3 How great is the problem? - Health Impact Assessment.1 Vol: 4 How dangerous is the problem? - Communicating the Risk.' Vol: 5 What we and our children need to know - Health & Hygiene Awareness.1 Still in preparation. This guide is available from: Director: Institute for Water Quality Studies Water Research Commission Department of Water Affairs & Forestry Private Bag X03 Private Bag X313 Geztna Pretoria 0031 0001 South Africa Tel: 012 808 0374 Tel: 012 330 0340 Fax: 012 808 0338 Fax: 012 331 2565 Disclaimer This report has been reviewed by the Water Research Commission (WRC) and approved for 1 publication. Approval does not signify that the contents necessarily reflect the views and policies of the WRC, nor does mention of trade names or commercial products constitute endorsement or f recommendation for use. -

STD (Sexually Transmitted Disease) Or STI (Sexually Transmitted Infection): Should We Choose? Janet Byron Anderson, Phd
STD (sexually transmitted disease) or STI (sexually transmitted infection): Should we choose? Janet Byron Anderson, PhD 1. Clinical foundation of the problem Another surprise: Documented incidence had shown Each of the terms—“sexual(ly)”, “transmitted”, “disease”, that the two routes of sexual transmission were male-to- and “infection”—is problematic independently, as this female and male-to-male. However, on 15 July 2016 the study will show. Moreover, co-occurring variants, used CDC reported the first case of suspected female-to-male as synonyms in English medical articles worldwide, ag- sexual transmission in New York City [4]. Since 2008, gravate the problem. The purpose of this study is to pro- then, the Zika virus has shown that it is now sexually pose a single term that can stimulate discussion about transmissible. “Transmissible” denotes a potential, and is whether the co-occurring variants are clinically and lin- distinct from “transmitted”, which denotes a reality. guistically justifiable—especially now, when STDs/STIs To say that Zika has proved to be transmissible through have become a global health problem, and public health sexual contact means that it can be transmitted through agencies in every country are scrambling to educate their sex but that it need not be (Zika remains a chiefly vec- citizens. [Until the presentation advances to the point tor-borne disease). However, cases reported since 2008 where the question posed in the title can be definitively document the reality of transmittedness through sexual answered, I’ll use the expression “STD/STI” or the terms contact. “illness(es)” and “condition(s)”.] The predisposing epidemiologic context will first be clar- This carefully differentiated language is used by the ified, for it sheds light on two of the problematic terms: European Centre for Disease Prevention and Control chiefly “transmitted” but also “sexual(ly)”. -

The Cursed Duet Today: TB and HIV-Coinfection
The cursed duet today: TB and HIV-coinfection Simon Tiberi#1, Anna Cristina C. Carvalho#2, Giorgia Sulis#3, Devan Vaghela#4, Adrian Rendon5, Fernanda C de Q Mello6, Ananna Rahman7, Nashaba Matin8, Alimuddin Zumla9, Emanuele Pontali10 # contributed equally Address of Correspondence: Division of Infection, Royal London Hospital, Barts Health NHS Trust. 80 Newark St. London E1 2ES, United Kingdom. Email: [email protected], Tel: +44 745001158 Affiliations 1. Division of Infection, Royal London Hospital, Barts Health NHS Trust, 80 Newark Street London E1 2ES, United Kingdom. Email: [email protected] 2. Laboratory of Innovations in Therapies, Education and Bioproducts, (LITEB), Oswaldo Cruz Institute (IOC), Fiocruz, Rio de Janeiro, Brazil. Email: [email protected] 3. University Department of Infectious and Tropical Diseases, World Health Organization Collaborating Centre for TB/HIV co-infection and TB elimination, University of Brescia, Brescia, Italy. Email: [email protected] 4. Department of Respiratory Medicine, Royal London Hospital, Barts Health NHS Trust, 80 Newark Street London E1 2ES, United Kingdom. Email: [email protected] 5. Centro de Investigación, Prevención y Tratamiento de Infecciones Respiratorias, Hospital Universitario de Monterrey, Monterrey, Nuevo León UANL, México. Email: [email protected] 6. Instituto de Doenças do Tórax (IDT)/Clementino Fraga Filho Hospital (CFFH), Federal University of Rio de Janeiro, Rua Professor Rodolpho Paulo Rocco, n° 255 - 1° Andar - Cidade Universitária - Ilha do Fundão, 21941-913, Rio De Janeiro, Brazil. Email: [email protected] 7. Department of Respiratory Medicine. Papworth Hospital NHS Foundation Trust, Papworth Everard, Cambridge, United Kingdom. Email: [email protected] 8. -

Colonization V. Infection
Colonization vs Infection Colonization • The presence of microorganisms in or on a host with growth and multiplication but without tissue invasion or damage • Understanding this concept is essential in the planning and implantation of epidemiological studies in a healthcare infection prevention and control program Infection v. Colonization • Confusing colonization with infection can lead to spurious associations that may lead to expensive, ineffective, and time‐ consuming interventions Multi Drug‐Resistant Organisms Management in Long Term Care • Colonization may become infection when changes in the host Facilities Workshop occur Louisiana Office of Public Health Healthcare‐Associated Infections Program Objectives Colonization: Definition By the end of the presentation, attendees will be able to: • Colonization: presence of a microorganism on/in a host, with • Define colonization growth and multiplication of the organism, but without • Differentiate colonization from infections interaction between host and organism (no clinical expression, no immune response). • Apply appropriate laboratory test by common LTC infectious • agents Carrier: individual which is colonized + more • • Understand the necessity of communicating infectious status Subclinical or unapparent infection: presence of upon patient transfer microorganism and interaction between host and microorganism (sub clinical response, immune response). Often the term colonization is applied for relationship host‐ agent in which the immune response is difficult to elicit. • Contamination: Presence of a microorganism on a body surface or an inanimate object. 1 Colonization vs Infection Spectrum: No Exposure ‐ Exposure ‐ Colonization ‐ Carrier Infection ‐ Disease A carrier is an individual that harbors a specific microorganism Host + Infectious agent What is “Exposed” ? in the absence of discernible clinical disease and serve as a No foothold: Exposed Means of transmission: potential source of infection. -

Occupational Immunity and Natural Vaccination
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/327185058 Occupational Immunity and Natural Vaccination Preprint · August 2018 DOI: 10.13140/RG.2.2.14987.34082 CITATIONS 0 2 authors, including: Steven A. Frank University of California, Irvine 235 PUBLICATIONS 10,574 CITATIONS SEE PROFILE All content following this page was uploaded by Steven A. Frank on 23 August 2018. The user has requested enhancement of the downloaded file. 1 Occupational Immunity and Natural Vaccination Robin M. Bush* & Steven A. Frank* *Department of Ecology and Evolutionary Biology, University of California, Irvine, CA 92697-2525 USA People who work with animals are frequently exposed to dangerous pathogens. Disease and subsequent immunity may result. Alternatively, occupational exposure to animals may lead to natural vaccination: the acquisition of immunity in the absence of overt disease. We use anthrax, Q fever, Campylobacter and influenza to illustrate aspects of dose, route and frequency of exposure that may be particularly favorable to natural vaccination. We then explore how exposure and immunity in those who work with animals provide clues about the epidemiology of emerging infectious diseases. Emerging infectious diseases arise from zoonotic pathogens that transmit from animals to humans. The most obvious recent zoonotic threat to human health comes from avian influenza. But the potential for zoonotic assault spans a wide array of pathogens including SARS, Ebola, anthrax, HIV, monkeypox, and the diverse bacteria of common farm animals that sometimes cause severe enteric or neural damage in humans. Zoonotic pathogens are also among the most commonly listed agents for use as bioterror weapons. -

Antibiotic Therapy for Subclinical Mastitis in Early Lactation
Copyright is owned by the Author of the thesis. Permission is given for a copy to be downloaded by an individual for the purpose of research and private study only. The thesis may not be reproduced elsewhere without the permission of the Author. ANTIBIOTIC THERAPY FOR SUBCLINICAL t�ASTITIS IN EARLY LACTATION ; EFFECTS ON INFECTION, SOMATIC CELL COUNTS AND ��ILK PR ODUCTION A THESIS PRESENTED IN PARTIAL FULFILMENT OF THE REQUIREMENTS FOR THE DEGREE OF MASTER OF AGRICULTURAL SCIENCE IN ANH·1AL SCIENCE AT MASSEY UNIVERSITY SAMSON MWAKASAGULE MWAKIPESILE ' .. '· 1982 ii ABSTRACT This study was conducted with the main purpose of investigating the effects of antibiotic therapy applied during lactation on the subclinically infected quarters in terms of bacterial infection , somatic cell count (S.C.C.) , milk yield and composition (i.e. milkfat and protein percentage). The main objectives involved were:- (1) To identify cows with high somatic cell counts (subclinical mastitis) in early lactation . (2) To identify the individual quarters with high somatic cell counts and bacterial infection . (3) To treat some of these high S.C.C. quarters with an antibiotic , and leave some of them untreated (control quarters) . (4) To measure the effects of treatment on S.C.C. , bacterial infection , yield and compo sition of milk . During weeks four to six of lactation 12 cows from a herd of 100 cows at No .3 Dairy Research & Development Unit, Massey University , were identified to have consistently high values for somatic cell counts . Milk samples were aseptically taken from the individual quarters of these cows and subjected to bacteriological tests. -
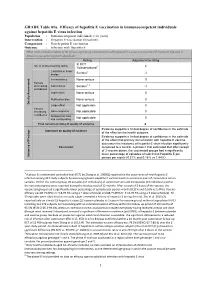
GRADE Table 01A. Efficacy of Hepatitis E Vaccination In
GRADE Table 01a. Efficacy of hepatitis E vaccination in immunocompetent individuals against hepatitis E virus infection Population : Immunocompetent individuals (>16 years) Intervention : Hepatitis E vaccination (Hecolin®) Comparison : Non-hepatitis E vaccination Outcome : Infection with Hepatitis E What is the scientific evidence of the efficacy of primary immunization with hepatitis E vaccine (versus control) to prevent hepatitis E disease in immunocompetent individuals? Rating Adjustment to rating 3/ RCT No. of studies/starting rating 1 4 1/observational Limitation in study Serious2 -1 design Inconsistency None serious 0 Factors decreasing Indirectness Serious3, 4 -1 confidence Imprecision None serious 0 Publication bias None serious 0 Quality Assessment Quality Large effect Not applicable 0 Factors increasing Dose-response Not applicable 0 confidence Antagonistic bias Not applicable 0 and confounding Final numerical rating of quality of evidence 2 Evidence supports a limited degree of confidence in the estimate Statement on quality of evidence of the effect on the health outcome. Evidence supports a limited degree of confidence in the estimate of the effect that primary immunization with hepatitis E vaccine decreases the incidence of hepatitis E virus infection significantly Conclusion compared to a control. A phase II trial estimated that after receipt of 3 vaccine doses, the vaccinated groups had a significantly lower percentage of episodes of subclinical Hepatitis E per Summary of Findings of Summary person per month (0.21% and 0.16% vs 1.44%) 1 A phase IIa randomized controlled trial (RCT) by Zhang et al. 2009(2) reported on the occurrence of new Hepatitis E infection among 457 study subjects by assessing IgG anti-Hepatitis E vaccine levels in successive pairs of consecutive serum samples. -

Assessment and Management of Infection in Alcoholic Hepatitis
Published online: 2019-08-01 THIEME 11 Assessment and Management of Infection in Alcoholic Hepatitis Nikhil Vergis, MRCP, PhD1 Stephen R. Atkinson, MRCP, PhD1 Mark R. Thursz, FRCP, MD1 1 Department of Surgery & Cancer, Division of Digestive Diseases, Address for correspondence Mark R. Thursz, FRCP, MD, Division of Imperial College London, London, United Kingdom Digestive Diseases, Department of Surgery & Cancer, Imperial College London, London, United Kingdom (e-mail: [email protected]). Semin Liver Dis 2020;40:11–19. Abstract Severe alcoholic hepatitis (SAH) is a condition characterized by jaundice and liver failure that develops after heavy and prolonged alcohol consumption. Infection frequently complicates the natural history of the disease and is independently associated with mortality. Objective recognition and recording of infection are therefore essential in Keywords the evaluation of therapeutic interventions and for antibiotic stewardship. This review ► alcoholic hepatitis will evaluate infections that complicate SAH at admission and beyond. Factors that ► infection associate with the development of infection will be identified and clinical and ► polymerase chain laboratory techniques available to identify infection will be discussed. Common reaction pathogens and frequently used antibiotics will be reviewed and recommendations ► systemic will be made for the management of infection for SAH patients. New techniques to inflammatory assess infection earlier and more precisely may improve diagnosis and treatmentof this response -

LEPTOSPIROSIS [Weil's Disease, Hemorrhagic Jaundice (Leptospira Icterohaemorrhagiae), Canicola Fever (L
LEPTOSPIROSIS [Weil's disease, Hemorrhagic jaundice (Leptospira icterohaemorrhagiae), canicola fever (L. canicola), dairy worker fever (L. hardjo)] SPECIES: rodents, domestic, and wild animals AGENT: Spirochete, Leptospira. Pathogenic leptospires belong to the species Leptospira interrogans, which is subdivided into more than 200 serovars. The main natural reservoirs for human infection vary with serovar: L. canicola in dogs, L. hardjo in cattle, L. pomona in swine, and L. icterohaemorrhagiae in rats. RESERVOIR AND INCIDENCE: Rats, mice, field moles, guinea pigs, gerbils, squirrels, rabbits, hamsters, reptiles, nonhuman primates, livestock, and dogs. In one study, 40 % of stray dogs were seropositive. Rats and mice are common animal hosts for L. ballum. Infection in mice is inapparent and can persist for the animal's lifetime. *Rodents are the only major animal species that can shed leptospires throughout their life-span without clinical manifestations. Active shedding by lab animals can go unrecognized until personnel handling the animals become clinically ill. TRANSMISSION: Handling infected animals, contaminating hands, or abrasions with urine, or aerosol exposure during cage cleaning are most common. The organism is often transmitted to humans by the urine of the reservoir host. The organism may also enter through minor skin lesions and probably via the conjunctiva. Many infections have followed bathing or swimming in infected waters. DISEASE IN ANIMALS: In cattle, fever and anorexia occur with rapid decline in milk yield and atypical mastitis. Pregnant cows abort with retention of the placenta. Also, mild jaundice and severe anemia occurs with enlarged and friable liver and swollen kidneys. In pigs subclinical infection is common, though it can cause abortion and birth of weak piglets.