Poročilo 2008
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Title Genomic Analysis of the Marine Hyperthermophilic Archaeon
Genomic analysis of the marine hyperthermophilic archaeon Title Aeropyrum( Digest_要約 ) Author(s) Daifuku, Takashi Citation 京都大学 Issue Date 2015-03-23 URL https://doi.org/10.14989/doctor.k19034 学位規則第9条第2項により要約公開; 許諾条件により本文 Right は2019-08-01に公開 Type Thesis or Dissertation Textversion ETD Kyoto University Summary The increasing number of genome sequences of archaea and bacteria leads to show their adaptation to different environmental conditions at the genomic level. Aeropyrum spp. are aerobic and hyperthermophilic archaea. A. camini was isolated from a deep-sea hydrothermal vent, and A. pernix was isolated from a coastal solfataric vent. In chapter 2, I compared the genomes of the two species to investigate the adaptation strategy in each habitat. Their shared genome features were a small genome size, a high GC content, and a large portion of orthologous genes (86 to 88%). The genomes also showed high synteny. These shared features may have been derived from the small number of mobile genetic elements and the lack of a RecBCD system, a recombinational enzyme complex. In addition, the specialized physiology (aerobic and hyperthermophilic) of Aeropyrum spp. may also contribute to the entire-genome similarity. Despite having stable genomes, interference of synteny occurred with two proviruses, A. pernix spindle-shaped virus 1 (APSV1) and A. pernix ovoid virus 1 (APOV1), and clustered regularly interspaced short palindromic repeat (CRISPR) elements. CRISPR spacer sequences observed in the A. camini showed significant matches with protospacers of the two proviruses found in the genome of A. pernix, indicating that A. camini interacted with viruses closely related to APSV1 and APOV1. Furthermore, a significant fraction of the nonorthologous genes (41 to 45%) were proviral genes or ORFans probably originating from viruses. -

Biological Diversity in the Patent System
Biological Diversity in the Patent System Paul Oldham1,2*, Stephen Hall1,3, Oscar Forero1,4 1 ESRC Centre for Economic and Social Aspects of Genomics (Cesagen), Lancaster University, Lancaster, United Kingdom, 2 Institute of Advanced Studies, United Nations University, Yokohama, Japan, 3 One World Analytics, Lancaster, United Kingdom, 4 Centre for Development, Environment and Policy, SOAS, University of London, London, United Kingdom Abstract Biological diversity in the patent system is an enduring focus of controversy but empirical analysis of the presence of biodiversity in the patent system has been limited. To address this problem we text mined 11 million patent documents for 6 million Latin species names from the Global Names Index (GNI) established by the Global Biodiversity Information Facility (GBIF) and Encyclopedia of Life (EOL). We identified 76,274 full Latin species names from 23,882 genera in 767,955 patent documents. 25,595 species appeared in the claims section of 136,880 patent documents. This reveals that human innovative activity involving biodiversity in the patent system focuses on approximately 4% of taxonomically described species and between 0.8–1% of predicted global species. In this article we identify the major features of the patent landscape for biological diversity by focusing on key areas including pharmaceuticals, neglected diseases, traditional medicines, genetic engineering, foods, biocides, marine genetic resources and Antarctica. We conclude that the narrow focus of human innovative activity and ownership of genetic resources is unlikely to be in the long term interest of humanity. We argue that a broader spectrum of biodiversity needs to be opened up to research and development based on the principles of equitable benefit-sharing, respect for the objectives of the Convention on Biological Diversity, human rights and ethics. -

Microbial Evolution and Diversity
PART V Microbial Evolution and Diversity This material cannot be copied, disseminated, or used in any way without the express written permission of the publisher. Copyright 2007 Sinauer Associates Inc. The objectives of this chapter are to: N Provide information on how bacteria are named and what is meant by a validly named species. N Discuss the classification of Bacteria and Archaea and the recent move toward an evolutionarily based, phylogenetic classification. N Describe the ways in which the Bacteria and Archaea are identified in the laboratory. This material cannot be copied, disseminated, or used in any way without the express written permission of the publisher. Copyright 2007 Sinauer Associates Inc. 17 Taxonomy of Bacteria and Archaea It’s just astounding to see how constant, how conserved, certain sequence motifs—proteins, genes—have been over enormous expanses of time. You can see sequence patterns that have per- sisted probably for over three billion years. That’s far longer than mountain ranges last, than continents retain their shape. —Carl Woese, 1997 (in Perry and Staley, Microbiology) his part of the book discusses the variety of microorganisms that exist on Earth and what is known about their characteris- Ttics and evolution. Most of the material pertains to the Bacteria and Archaea because there is a special chapter dedicated to eukaryotic microorganisms. Therefore, this first chapter discusses how the Bacte- ria and Archaea are named and classified and is followed by several chapters (Chapters 18–22) that discuss the properties and diversity of the Bacteria and Archaea. When scientists encounter a large number of related items—such as the chemical elements, plants, or animals—they characterize, name, and organize them into groups. -

Archaeal Viruses and Bacteriophages: Comparisons and Contrasts
Review Archaeal viruses and bacteriophages: comparisons and contrasts Maija K. Pietila¨ , Tatiana A. Demina, Nina S. Atanasova, Hanna M. Oksanen, and Dennis H. Bamford Institute of Biotechnology and Department of Biosciences, P.O. Box 56, Viikinkaari 5, 00014 University of Helsinki, Helsinki, Finland Isolated archaeal viruses comprise only a few percent of Euryarchaeaota [9,10]. Archaea have also been cultivated all known prokaryotic viruses. Thus, the study of viruses from moderate environments such as seawater and soil. infecting archaea is still in its early stages. Here we Consequently, an additional phylum, Thaumarchaeota, summarize the most recent discoveries of archaeal vi- has been formed to contain mesophilic and thermophilic ruses utilizing a virion-centered view. We describe the ammonia-oxidizing archaea [11]. However, all known ar- known archaeal virion morphotypes and compare them chaeal viruses infect extremophiles – mainly hyperther- to the bacterial counterparts, if such exist. Viruses infect- mophiles belonging to the crenarchaeal genera Sulfolobus ing archaea are morphologically diverse and present and Acidianus or halophiles of the euryarchaeal genera some unique morphotypes. Although limited in isolate Haloarcula, Halorubrum, and Halobacterium [6,7]. Even number, archaeal viruses reveal new insights into the though bacteria are also found in diverse extreme habitats viral world, such as deep evolutionary relationships such as hypersaline lakes, archaea typically dominate at between viruses that infect hosts from all three domains extreme salinities, based on both cultivation-dependent of life. and -independent studies [6,12–15]. Consequently, archae- al viruses do the same in hypersaline environments. About Discovery of archaeal viruses 50 prokaryotic haloviruses were recently isolated from All cellular organisms are susceptible to viral infections, nine globally distant locations, and only four of them which makes viruses a major evolutionary force shaping infected bacteria [6,16]. -

Differences in Lateral Gene Transfer in Hypersaline Versus Thermal Environments Matthew E Rhodes1*, John R Spear2, Aharon Oren3 and Christopher H House1
Rhodes et al. BMC Evolutionary Biology 2011, 11:199 http://www.biomedcentral.com/1471-2148/11/199 RESEARCH ARTICLE Open Access Differences in lateral gene transfer in hypersaline versus thermal environments Matthew E Rhodes1*, John R Spear2, Aharon Oren3 and Christopher H House1 Abstract Background: The role of lateral gene transfer (LGT) in the evolution of microorganisms is only beginning to be understood. While most LGT events occur between closely related individuals, inter-phylum and inter-domain LGT events are not uncommon. These distant transfer events offer potentially greater fitness advantages and it is for this reason that these “long distance” LGT events may have significantly impacted the evolution of microbes. One mechanism driving distant LGT events is microbial transformation. Theoretically, transformative events can occur between any two species provided that the DNA of one enters the habitat of the other. Two categories of microorganisms that are well-known for LGT are the thermophiles and halophiles. Results: We identified potential inter-class LGT events into both a thermophilic class of Archaea (Thermoprotei) and a halophilic class of Archaea (Halobacteria). We then categorized these LGT genes as originating in thermophiles and halophiles respectively. While more than 68% of transfer events into Thermoprotei taxa originated in other thermophiles, less than 11% of transfer events into Halobacteria taxa originated in other halophiles. Conclusions: Our results suggest that there is a fundamental difference between LGT in thermophiles and halophiles. We theorize that the difference lies in the different natures of the environments. While DNA degrades rapidly in thermal environments due to temperature-driven denaturization, hypersaline environments are adept at preserving DNA. -

Title Genomic Analysis of the Marine Hyperthermophilic Archaeon
Genomic analysis of the marine hyperthermophilic archaeon Title Aeropyrum( Dissertation_全文 ) Author(s) Daifuku, Takashi Citation 京都大学 Issue Date 2015-03-23 URL https://doi.org/10.14989/doctor.k19034 学位規則第9条第2項により要約公開; 許諾条件により本文 Right は2019-08-01に公開 Type Thesis or Dissertation Textversion ETD Kyoto University 1. General introduction Chapter 1 General introduction Gene repertoires and genome organizations differ between closely related microbial organisms depending on the ecological characteristics of each habitat (Cohan and Koeppel 2008). The cyanobacterial Prochlorococcus spp. account for a significant fraction of primary production in the ocean (Goericke and Welschmeyer 1993) and show physiological features relevant to the different ecological niches within a stratified oceanic water column (Moore et al. 1998; West et al. 2001). The whole-genomic comparisons of the Prochlorococcus spp. strains show gross signatures according to this niche differentiation (Rocap et al. 2003). Alpha-proteobacterium Pelagibacter ubique which belongs to the SAR11 clade in the phylogenetic tree based on the 16S rRNA gene is the most abundant microorganism in the ocean (Morris et al. 2002). The genomes of the SAR11 isolates are highly conserved in the core genes that are common to all strains (Medini et al. 2005) and show synteny (the conservation of DNA sequence and gene order) (Bentley and Parkhill 2004). However, variations exist among genes for phosphorus metabolism, glycolysis, and C1 metabolism, suggesting that adaptive specialization in nutrient resource utilization is important for niche partitioning (Grote et al. 2012). This adaptation at the genomic level was also observed in archaea. The members of the genus Pyrococcus are anaerobic and hyperthermophilic archaea (Fiala and Stetter 1 1. -

Reading Phylogenetic Trees: a Quick Review (Adapted from Evolution.Berkeley.Edu)
Biological Trees Gloria Rendon SC11 Education June, 2011 Biological trees • Biological trees are used for the purpose of classification, i.e. grouping and categorization of organisms by biological type such as genus or species. Types of Biological trees • Taxonomy trees, like the one hosted at NCBI, are hierarchies; thus classification is determined by position or rank within the hierarchy. It goes from kingdom to species. • Phylogenetic trees represent evolutionary relationships, or genealogy, among species. Nowadays, these trees are usually constructed by comparing 16s/18s ribosomal RNA. • Gene trees represent evolutionary relationships of a particular biological molecule (gene or protein product) among species. They may or may not match the species genealogy. Examples: hemoglobin tree, kinase tree, etc. TAXONOMY TREES Exercise 1: Exploring the Species Tree at NCBI •There exist many taxonomies. •In this exercise, we will examine the taxonomy at NCBI. •NCBI has a taxonomy database where each category in the tree (from the root to the species level) has a unique identifier called taxid. •The lineage of a species is the full path you take in that tree from the root to the point where that species is located. •The (NCBI) taxonomy common tree is therefore the tree that results from adding together the full lineages of each species in a particular list of your choice. Exercise 1: Exploring the Species Tree at NCBI • Open a web browser on NCBI’s Taxonomy page http://www.ncbi.nlm.n ih.gov/Taxonomy/ • Click on each one of the names here to look up the taxonomy id (taxid) of each one of the five categories of the taxonomy browser: Archaea, bacteria, Eukaryotes, Viroids and Viruses. -

Pyrolobus Fumarii Type Strain (1A)
Lawrence Berkeley National Laboratory Recent Work Title Complete genome sequence of the hyperthermophilic chemolithoautotroph Pyrolobus fumarii type strain (1A). Permalink https://escholarship.org/uc/item/89r1s0xt Journal Standards in genomic sciences, 4(3) ISSN 1944-3277 Authors Anderson, Iain Göker, Markus Nolan, Matt et al. Publication Date 2011-07-01 DOI 10.4056/sigs.2014648 Peer reviewed eScholarship.org Powered by the California Digital Library University of California Standards in Genomic Sciences (2011) 4:381-392 DOI:10.4056/sigs.2014648 Complete genome sequence of the hyperthermophilic chemolithoautotroph Pyrolobus fumarii type strain (1AT) Iain Anderson1, Markus Göker2, Matt Nolan1, Susan Lucas1, Nancy Hammon1, Shweta Deshpande1, Jan-Fang Cheng1, Roxanne Tapia1,3, Cliff Han1,3, Lynne Goodwin1,3, Sam Pitluck1, Marcel Huntemann1, Konstantinos Liolios1, Natalia Ivanova1, Ioanna Pagani1, Konstantinos Mavromatis1, Galina Ovchinikova1, Amrita Pati1, Amy Chen4, Krishna Pala- niappan4, Miriam Land1,5, Loren Hauser1,5, Evelyne-Marie Brambilla2, Harald Huber6, Montri Yasawong7, Manfred Rohde7, Stefan Spring2, Birte Abt2, Johannes Sikorski2, Reinhard Wirth6, John C. Detter1,3, Tanja Woyke1, James Bristow1, Jonathan A. Eisen1,8, Victor Markowitz4, Philip Hugenholtz1,9, Nikos C. Kyrpides1, Hans-Peter Klenk2, and Alla Lapidus1* 1 DOE Joint Genome Institute, Walnut Creek, California, USA 2 DSMZ - German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany 3 Los Alamos National Laboratory, Bioscience Division, Los Alamos, -
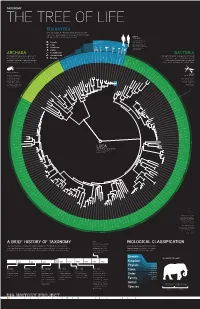
A Brief History of Taxonomy Biological Classification
TAXONOMY THE TREE OF LIFE EUKARYOTA This domain includes all of the plants, animals, and fungi, and some single-celled organisms. Eukaryotes are distinguished by their complex cells, which contain a membrane-enclosed nucleus. Humans Homo sapiens The creatures most familiar to us, Our species, primates in the animals, are members of the Animalia kingdom of the Animalia same kingdom. Eukaryota, is thought to have Fungi Mosquito Red first evolved in Africa about Pufferfish Junglefowl Roundworm Mouse 200,000 years ago. Genetically, Amoebozoa Chimpanzee our closest living relative Plantae is the chimpanzee. Archaeplastida Schizosaccharomyces pombe ARCHAEA Saccharomyces cerevisiae BACTERIA Caenorhabditis briggsae Caenorhabditis elegans Eremothecium gossypii These single-celled prokaryotic organisms often Chromalveolata Dictyostelium discoideum These single-celled prokaryotic organisms were among Cyanidioschyzon merolae live in extreme environmental conditions. Once Excavata Arabidopsis thaliana the first life forms to appear on Earth. Often spherical, Plasmodium falciparum considered to be Bacteria, these microorganisms Cryptosporidium hominis rod-like, or spiral in shape, these microorganisms Thalassiosira pseudonana Oryza sativa Anopheles gambiae Drosophila melanogaster Takifugu rubripes Danio rerio are now recognized as a separate domain of life. Gallus gallus function without a membrane-enclosed cell nucleus. Rattus norvegicus Mus musculus Methanococcus jannaschii Leishmania major Homo sapiens Pan troglodytes Methanococcus maripaludi Thermoanaerobacter -

Chapter 20974
Genome Replication of Bacterial and Archaeal Viruses Česlovas Venclovas, Vilnius University, Vilnius, Lithuania r 2019 Elsevier Inc. All rights reserved. Glossary RNA-primed DNA replication Conventional DNA Negative sense ( À ) strand A negative-sense DNA or RNA replication used by all cellular organisms whereby a strand has a nucleotide sequence complementary to the primase synthesizes a short RNA primer with a free 3′-OH messenger RNA and cannot be directly translated into protein. group which is subsequently elongated by a DNA Positive sense (+) strand A positive sense DNA or RNA polymerase. strand has a nucleotide sequence, which is the same as that Rolling-circle DNA replication DNA replication whereby of the messenger RNA, and the RNA version of this sequence the replication initiation protein creates a nick in the circular is directly translatable into protein. double-stranded DNA and becomes covalently attached to Protein-primed DNA replication DNA replication whereby the 5′ end of the nicked strand. The free 3′-OH group at the a DNA polymerase uses the 3′-OH group provided by the nick site is then used by the DNA polymerase to synthesize specialized protein as a primer to synthesize a new DNA strand. the new strand. Genomes of Prokaryotic Viruses At present, all identified archaeal viruses have either double-stranded (ds) or single-stranded (ss) DNA genomes. Although metagenomic analyzes suggested the existence of archaeal viruses with RNA genomes, this finding remains to be substantiated. Bacterial viruses, also refered to as bacteriophages or phages for short, have either DNA or RNA genomes, including circular ssDNA, circular or linear dsDNA, linear positive-sense (+)ssRNA or segmented dsRNA (Table 1). -

Biotechnology of Archaea- Costanzo Bertoldo and Garabed Antranikian
BIOTECHNOLOGY– Vol. IX – Biotechnology Of Archaea- Costanzo Bertoldo and Garabed Antranikian BIOTECHNOLOGY OF ARCHAEA Costanzo Bertoldo and Garabed Antranikian Technical University Hamburg-Harburg, Germany Keywords: Archaea, extremophiles, enzymes Contents 1. Introduction 2. Cultivation of Extremophilic Archaea 3. Molecular Basis of Heat Resistance 4. Screening Strategies for the Detection of Novel Enzymes from Archaea 5. Starch Processing Enzymes 6. Cellulose and Hemicellulose Hydrolyzing Enzymes 7. Chitin Degradation 8. Proteolytic Enzymes 9. Alcohol Dehydrogenases and Esterases 10. DNA Processing Enzymes 11. Archaeal Inteins 12. Conclusions Glossary Bibliography Biographical Sketches Summary Archaea are unique microorganisms that are adapted to survive in ecological niches such as high temperatures, extremes of pH, high salt concentrations and high pressure. They produce novel organic compounds and stable biocatalysts that function under extreme conditions comparable to those prevailing in various industrial processes. Some of the enzymes from Archaea have already been purified and their genes successfully cloned in mesophilic hosts. Enzymes such as amylases, pullulanases, cyclodextrin glycosyltransferases, cellulases, xylanases, chitinases, proteases, alcohol dehydrogenase,UNESCO esterases, and DNA-modifying – enzymesEOLSS are of potential use in various biotechnological processes including in the food, chemical and pharmaceutical industries. 1. Introduction SAMPLE CHAPTERS The industrial application of biocatalysts began in 1915 with the introduction of the first detergent enzyme by Dr. Röhm. Since that time enzymes have found wider application in various industrial processes and production (see Enzyme Production). The most important fields of enzyme application are nutrition, pharmaceuticals, diagnostics, detergents, textile and leather industries. There are more than 3000 enzymes known to date that catalyze different biochemical reactions among the estimated total of 7000; only 100 enzymes are being used industrially. -

Modified Mevalonate Pathway of the Archaeon Aeropyrum Pernix Proceeds Via Trans- Anhydromevalonate 5-Phosphate
Modified mevalonate pathway of the archaeon Aeropyrum pernix proceeds via trans- anhydromevalonate 5-phosphate Hajime Hayakawaa, Kento Motoyamaa, Fumiaki Sobuea, Tomokazu Itoa, Hiroshi Kawaideb, Tohru Yoshimuraa, and Hisashi Hemmia,1 aDepartment of Applied Molecular Bioscience, Graduate School of Bioagricultural Sciences, Nagoya University, Nagoya, 464-8601 Aichi, Japan; and bInstitute of Symbiotic Science and Technology, Tokyo University of Agriculture and Technology, Fuchu, 183-8509 Tokyo, Japan Edited by C. Dale Poulter, University of Utah, Salt Lake City, UT, and approved August 23, 2018 (received for review May 28, 2018) The modified mevalonate pathway is believed to be the upstream MVA pathway exists in almost all eukaryotes and in certain forms biosynthetic route for isoprenoids in general archaea. The partially of bacteria, such as lactic acid bacteria, whereas the vast majority identified pathway has been proposed to explain a mystery of bacteria utilize the methylerythritol phosphate (MEP) pathway surrounding the lack of phosphomevalonate kinase and diphospho- that proceeds through completely different intermediates from mevalonate decarboxylase by the discovery of a conserved enzyme, those in the MVA pathway. isopentenyl phosphate kinase. Phosphomevalonate decarboxylase The “modified” MVA pathway was first proposed in 2006 by was considered to be the missing link that would fill the vacancy in Grochowski et al. (4) based on the discovery of a new enzyme, the pathway between mevalonate 5-phosphate and isopentenyl isopentenyl phosphate kinase (IPK), and on data from compar- phosphate. This enzyme was recently discovered from haloarchaea ative analyses of archaeal genomes. For archaea, which do not and certain Chroloflexi bacteria, but their enzymes are close homo- possess the MEP pathway, the MVA pathway is requisite for the logs of diphosphomevalonate decarboxylase, which are absent in biosynthesis of specific membrane lipids and other isoprenoids, most archaea.