Genome Sequence and Gene Compaction of the Eukaryote
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Emerging Regulatory Roles of Mitochondrial Sirtuins on Pyruvate Dehydrogenase Complex and the Related Metabolic Diseases: Review
Biomedical Research and Therapy, 7(2):3645-3658 Open Access Full Text Article Review Emerging regulatory roles of mitochondrial sirtuins on pyruvate dehydrogenase complex and the related metabolic diseases: Review Abolfazl Nasiri1,2, Masoud Sadeghi3, Asad Vaisi-Raygani2,4, Sara Kiani3, Zahra Aghelan1,2, Reza Khodarahmi3,5,* ABSTRACT The pyruvate dehydrogenase complex (PDC) is a multi-enzyme complex of the mitochondria that provides a link between glycolysis and the Krebs cycle. PDC plays an essential role in producing Use your smartphone to scan this acetyl-CoA from glucose and the regulation of fuel consumption. In general, PDC enzyme is reg- QR code and download this article ulated in two different ways, end-product inhibition and posttranslational modifications (moreex- tensive phosphorylation and dephosphorylation subunit E1). Posttranslational modifications of this enzyme are regulated by various factors. Sirtuins are the class III of histone deacylatases that catalyze 1Students Research Committee, protein posttranslational modifications, including deacetylation, adenosine diphosphate ribosyla- Kermanshah University of Medical tion, and deacylation. Sirt3, Sirt4, and Sirt5 are mitochondrial sirtuins that control the posttrans- Sciences, Kermanshah, Iran lational modifications of mitochondrial protein. Considering the comprehensive role of sirtuins in post-translational modifications and regulation of metabolic processes, the aim of this review isto 2 Department of Clinical Biochemistry, explore the role of mitochondrial sirtuins in the regulation of the PDC. PDC deficiency is a com- School of Medicine, Kermanshah mon metabolic disorder that causes pyruvate to be converted to lactate and alanine rather than to University of Medical Sciences, Kermanshah, Iran acetyl-CoA. because this enzyme is in the gateway of complete oxidation, glucose products enter- ing the Krebs cycle and resulting in physiological and structural changes in the organs. -

Alternatives in Molecular Diagnostics of Encephalitozoon and Enterocytozoon Infections
Journal of Fungi Review Alternatives in Molecular Diagnostics of Encephalitozoon and Enterocytozoon Infections Alexandra Valenˇcáková * and Monika Suˇcik Department of Biology and Genetics, University of Veterinary Medicine and Pharmacy, Komenského 73, 04181 Košice, Slovakia; [email protected] * Correspondence: [email protected] Received: 15 June 2020; Accepted: 20 July 2020; Published: 22 July 2020 Abstract: Microsporidia are obligate intracellular pathogens that are currently considered to be most directly aligned with fungi. These fungal-related microbes cause infections in every major group of animals, both vertebrate and invertebrate, and more recently, because of AIDS, they have been identified as significant opportunistic parasites in man. The Microsporidia are ubiquitous parasites in the animal kingdom but, until recently, they have maintained relative anonymity because of the specialized nature of pathology researchers. Diagnosis of microsporidia infection from stool examination is possible and has replaced biopsy as the initial diagnostic procedure in many laboratories. These staining techniques can be difficult, however, due to the small size of the spores. The specific identification of microsporidian species has classically depended on ultrastructural examination. With the cloning of the rRNA genes from the human pathogenic microsporidia it has been possible to apply polymerase chain reaction (PCR) techniques for the diagnosis of microsporidial infection at the species and genotype level. The absence of genetic techniques for manipulating microsporidia and their complicated diagnosis hampered research. This study should provide basic insights into the development of diagnostics and the pitfalls of molecular identification of these ubiquitous intracellular pathogens that can be integrated into studies aimed at treating or controlling microsporidiosis. Keywords: Encephalitozoon spp.; Enterocytozoonbieneusi; diagnosis; molecular diagnosis; primers 1. -

Elevation of Cardiac Glycolysis Reduces Pyruvate Dehydrogenase but Increases Glucose Oxidation
University of Louisville ThinkIR: The University of Louisville's Institutional Repository Electronic Theses and Dissertations 5-2011 Elevation of cardiac glycolysis reduces pyruvate dehydrogenase but increases glucose oxidation. Qianwen Wang University of Louisville Follow this and additional works at: https://ir.library.louisville.edu/etd Recommended Citation Wang, Qianwen, "Elevation of cardiac glycolysis reduces pyruvate dehydrogenase but increases glucose oxidation." (2011). Electronic Theses and Dissertations. Paper 1519. https://doi.org/10.18297/etd/1519 This Doctoral Dissertation is brought to you for free and open access by ThinkIR: The University of Louisville's Institutional Repository. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of ThinkIR: The University of Louisville's Institutional Repository. This title appears here courtesy of the author, who has retained all other copyrights. For more information, please contact [email protected]. ELEVATION OF CARDIAC GLYCOLYSIS REDUCES PYRUVATE DEHYDROGENASE BUT INCREASES GLUCOSE OXIDATION By Qianwen Wang B.S., Guangdong Medical College of China, 1999 M.S., University of Louisville, 2006 A Dissertaion Submitted to the Faculty of the Graduate School of the University of Louisville In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Department of Physiology and Biophysics University of Louisville, School of Medicine Louisville, Kentucky May 2011 ELEVATION OF CARDIAC GLYCOLYSIS REDUCES PYRUVATE DEHYDROGENASE BUT INCREASES GLUCOSE OXIDATION By Qianwen Wang B.S., Guangdong Medical College of China, 1999 M.S., University of Louisville, 2006 A Dissertation Approved on March 28, 2011 by the following Dissertation Committee: Dissertation Director-Paul N. Epstein, Ph.D. William B. -

The Apicoplast: a Review of the Derived Plastid of Apicomplexan Parasites
Curr. Issues Mol. Biol. 7: 57-80. Online journalThe Apicoplastat www.cimb.org 57 The Apicoplast: A Review of the Derived Plastid of Apicomplexan Parasites Ross F. Waller1 and Geoffrey I. McFadden2,* way to apicoplast discovery with studies of extra- chromosomal DNAs recovered from isopycnic density 1Botany, University of British Columbia, 3529-6270 gradient fractionation of total Plasmodium DNA. This University Boulevard, Vancouver, BC, V6T 1Z4, Canada group recovered two DNA forms; one a 6kb tandemly 2Plant Cell Biology Research Centre, Botany, University repeated element that was later identifed as the of Melbourne, 3010, Australia mitochondrial genome, and a second, 35kb circle that was supposed to represent the DNA circles previously observed by microscopists (Wilson et al., 1996b; Wilson Abstract and Williamson, 1997). This molecule was also thought The apicoplast is a plastid organelle, homologous to to be mitochondrial DNA, and early sequence data of chloroplasts of plants, that is found in apicomplexan eubacterial-like rRNA genes supported this organellar parasites such as the causative agents of Malaria conclusion. However, as the sequencing effort continued Plasmodium spp. It occurs throughout the Apicomplexa a new conclusion, that was originally embraced with and is an ancient feature of this group acquired by the some awkwardness (“Have malaria parasites three process of endosymbiosis. Like plant chloroplasts, genomes?”, Wilson et al., 1991), began to emerge. apicoplasts are semi-autonomous with their own genome Gradually, evermore convincing character traits of a and expression machinery. In addition, apicoplasts import plastid genome were uncovered, and strong parallels numerous proteins encoded by nuclear genes. These with plastid genomes from non-photosynthetic plants nuclear genes largely derive from the endosymbiont (Epifagus virginiana) and algae (Astasia longa) became through a process of intracellular gene relocation. -

Cellular Respiration
Cellular Respiration Glucose is the preferred carbohydrate of cells. In solution, it can change from a linear chain to a ring. Energy is stored in the bonds of the carbohydrates. Breaking these bonds releases that energy. Crushing sugar crystals creates tiny electrical fields that give off invisible ultraviolet light. The wintergreen chemical (methyl salicylate) gets excited by these excited electrons and fluoresces in a visible blue wavelength. This phenomenon is called triboluminescence. Contents 1 Glycolysis 2 Fermentation 3 The Preparatory Reaction 4 Mitochondria 5 Aerobic Respiration 6 Metabolic Pool Glycolysis Glucose is the preferred carbohydrate of cells. Glycolysis (glyco – sugar; lysis – splitting) is a universal process of all cells that occurs in the cytosol whereby the glucose (a 6-carbon sugar) is split into two pyruvate (a 3-carbon molecule) molecules to generate ATP and reduced NADH. ATP (adenosine triphosphate) is the energy currency of the cell that stores chemical energy in 3 high energy phosphate bonds. NADH (reduced nicotinamide adenine dinucleotide) is a high energy electron carrier that acts as a coenzyme in reactions and as a rechargeable battery of sorts. The uncharged state that is not carrying high energy electrons is called NAD+. CC-BY-NC-SA | Jeremy Seto | New York City College of Technology | 1 Cellular Respiration Glycolysis is the splitting of glucose into 2 pyruvate molecules to generate 2 NADH and 2ATP molecules. ATP contains 3 high energy phosphates and acts as cellular energy currency. NADH is the reduced form of NAD+. The High energy electrons associated with the reduced form come with a H atom. -

CELLULAR RESPIRATION Applied Medical Chemistry II Course
CELLULAR RESPIRATION Applied Medical Chemistry II Course Aim of the lecture This lecture discuss the steps of cellular respiration and its regulation Maye M. Ragab Super Visor / Dr. Neveen Introduction individual enzymic reactions were analyzed in an effort to explain the mechanisms of catalysis. However, in cells, these reactions rarely occur in isolation, but rather are organized into multistep sequences called pathways). In a pathway, the product of one reaction serves as the substrate of the subsequent reaction. Different pathways can also intersect, forming an integrated and purposeful network of chemical reactions. These are collectively called metabolism, which is the sum of all the chemical changes occurring in a cell, a tissue, or the body. Most pathways can be classified as either catabolic (degradative) or anabolic (synthetic). Anabolic pathways • form complex end products from simple precursors, for example, the synthesis of the polysaccharide, glycogen, from glucose. • Anabolic reactions require energy (are endergonic), which is generally provided by the breakdown of ATP to (ADP) and inorganic phosphate (Pi). • Anabolic reactions often involve chemical reductions in which the reducing power is most frequently provided by the electron donor NADPH . Catabolic reactions • break down complex molecules, such as proteins, polysaccharides, and lipids, to a few simple molecules, for example, CO2, NH3 (ammonia), and water. • Catabolic reactions serve to capture chemical energy in the form of (ATP) from the degradation of energy-rich fuel molecules. Energy generation by degradation of complex molecules occurs in three stages as in the following figure : [Note: Catabolic pathways are typically oxidative, and require coenzymes such as NAD+.] [Note: Pathways that regenerate a component are called cycles.] 1 2 Overview Of Glycolysis : • The glycolytic pathway is employed by all tissues for the breakdown of glucose to provide energy (in the form of ATP) and intermediates for other metabolic pathways. -

The PEP–Pyruvate–Oxaloacetate Node As the Switch Point for Carbon flux Distribution in Bacteria
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by RERO DOC Digital Library FEMS Microbiology Reviews 29 (2005) 765–794 www.fems-microbiology.org The PEP–pyruvate–oxaloacetate node as the switch point for carbon flux distribution in bacteria Uwe Sauer a, Bernhard J. Eikmanns b,* a Institute of Biotechnology, ETH Zu¨rich, 8093 Zu¨rich, Switzerland b Department of Microbiology and Biotechnology, University of Ulm, 89069 Ulm, Germany Received 18 August 2004; received in revised form 27 October 2004; accepted 1 November 2004 First published online 28 November 2004 We dedicate this paper to Rudolf K. Thauer, Director of the Max-Planck-Institute for Terrestrial Microbiology in Marburg, Germany, on the occasion of his 65th birthday Abstract In many organisms, metabolite interconversion at the phosphoenolpyruvate (PEP)–pyruvate–oxaloacetate node involves a struc- turally entangled set of reactions that interconnects the major pathways of carbon metabolism and thus, is responsible for the dis- tribution of the carbon flux among catabolism, anabolism and energy supply of the cell. While sugar catabolism proceeds mainly via oxidative or non-oxidative decarboxylation of pyruvate to acetyl-CoA, anaplerosis and the initial steps of gluconeogenesis are accomplished by C3- (PEP- and/or pyruvate-) carboxylation and C4- (oxaloacetate- and/or malate-) decarboxylation, respectively. In contrast to the relatively uniform central metabolic pathways in bacteria, the set of enzymes at the PEP–pyruvate–oxaloacetate node represents a surprising diversity of reactions. Variable combinations are used in different bacteria and the question of the sig- nificance of all these reactions for growth and for biotechnological fermentation processes arises. -

In Vivo Assessment of Pyruvate Dehydrogenase Flux in the Heart Using Hyperpolarized Carbon-13 Magnetic Resonance
In vivo assessment of pyruvate dehydrogenase flux in the heart using hyperpolarized carbon-13 magnetic resonance Marie A. Schroeder, Lowri E. Cochlin, Lisa C. Heather, Kieran Clarke, George K. Radda, and Damian J. Tyler* Cardiac Metabolism Research Group, Department of Physiology, Anatomy, and Genetics, University of Oxford, Oxford OX1 3PT, United Kingdom Communicated by Robert G. Shulman, Yale University, New Haven, CT, June 19, 2008 (received for review January 15, 2008) The advent of hyperpolarized 13C magnetic resonance (MR) has substrate composition can alter the phosphorylation state and provided new potential for the real-time visualization of in vivo thus activity of the PDH enzyme complex (7). Elevated insulin metabolic processes. The aim of this work was to use hyperpolar- concentrations are thought to stimulate PDP acutely, thereby ized [1-13C]pyruvate as a metabolic tracer to assess noninvasively dephosphorylating and activating PDH (9). The relative activi- the flux through the mitochondrial enzyme complex pyruvate ties of PDK and PDP determine the proportion of PDH that is dehydrogenase (PDH) in the rat heart, by measuring the produc- in the active dephosphorylated form. ؊ 13 tion of bicarbonate (H CO3 ), a byproduct of the PDH-catalyzed The link between PDH regulation and disease has made the conversion of [1-13C]pyruvate to acetyl-CoA. By noninvasively assessment of substrate selection and PDH activity important ؊ 13 observing a 74% decrease in H CO3 production in fasted rats benchmarks in the study of heart disease. To date, measurement compared with fed controls, we have demonstrated that hyper- of cardiac PDH activity has been performed with invasive 13 polarized C MR is sensitive to physiological perturbations in PDH biochemical assays, whereas substrate selection has been mon- 13 flux. -

Immunohistochemical Detection of Encephalitozoon Cuniculi in Ocular Structures of Immunocompetent Rabbits
animals Article Immunohistochemical Detection of Encephalitozoon cuniculi in Ocular Structures of Immunocompetent Rabbits Edita Jeklová 1, Lenka Levá 1, Vladimír Kummer 1, Vladimír Jekl 2 and Martin Faldyna 1,* 1 Veterinary Research Institute, Hudcova 296/70, 621 00 Brno, Czech Republic; [email protected] (E.J.); [email protected] (L.L.); [email protected] (V.K.) 2 Jekl & Hauptman Veterinary Clinic, Mojmírovo nám. 3105/6a, 612 00 Brno, Czech Republic; [email protected] * Correspondence: [email protected] Received: 11 October 2019; Accepted: 14 November 2019; Published: 18 November 2019 Simple Summary: Encephalitozoonosis is a common infectious disease widely spread among rabbits. Its causative agent, Encephalitozoon cuniculi, is considered to be transmissible to humans. In rabbits, clinical signs include discoordination, head tilt, excessive water intake, excessive urination and cataracts. This study investigates, for the first time, whether the E. cuniculi organism can be detected in ocular structures in healthy adult rabbits after experimental oral infection using immunohistochemistry—detection of the organism in the tissue using a specific staining method. In infected animals, E. cuniculi spores were detected in many ophthalmic structures (periocular connective tissue, sclera, cornea, choroidea, iris, retina and lens) as early as 2 weeks after infection. There were no signs of inflammatory lesions in any of the ocular tissues examined at 2, 4, 6 and 8 weeks after infection. E. cuniculi was also detected in the lens of adult rabbits, which indicates that ways of lens infection other than intrauterine and haematogenic are possible. This information can help to understand E. cuniculi dissemination to various ocular tissues structures after oral infection. -

Plastid-Targeting Peptides from the Chlorarachniophyte Bigelowiella Natans
J. Eukaryot. Microbiol., 51(5), 2004 pp. 529±535 q 2004 by the Society of Protozoologists Plastid-Targeting Peptides from the Chlorarachniophyte Bigelowiella natans MATTHEW B. ROGERS,a JOHN M. ARCHIBALD,a,1 MATTHEW A. FIELD,a CATHERINE LI,b BORIS STRIEPENb and PATRICK J. KEELINGa aCanadian Institute for Advanced Research, Department of Botany, University of British Columbia, 3529-6270 University Boulevard, Vancouver, BC, V6T 1Z4, Canada, and bCenter for Tropical & Emerging Global Diseases & Department of Cellular Biology, University of Georgia, 724 Biological Sciences Building, Athens, Georgia 30602, USA ABSTRACT. Chlorarachniophytes are marine amoebo¯agellate protists that have acquired their plastid (chloroplast) through secondary endosymbiosis with a green alga. Like other algae, most of the proteins necessary for plastid function are encoded in the nuclear genome of the secondary host. These proteins are targeted to the organelle using a bipartite leader sequence consisting of a signal peptide (allowing entry in to the endomembrane system) and a chloroplast transit peptide (for transport across the chloroplast envelope mem- branes). We have examined the leader sequences from 45 full-length predicted plastid-targeted proteins from the chlorarachniophyte Bigelowiella natans with the goal of understanding important features of these sequences and possible conserved motifs. The chemical characteristics of these sequences were compared with a set of 10 B. natans endomembrane-targeted proteins and 38 cytosolic or nuclear proteins, which show that the signal peptides are similar to those of most other eukaryotes, while the transit peptides differ from those of other algae in some characteristics. Consistent with this, the leader sequence from one B. -
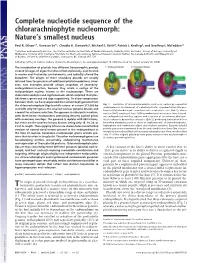
Complete Nucleotide Sequence of the Chlorarachniophyte Nucleomorph: Nature’S Smallest Nucleus
Complete nucleotide sequence of the chlorarachniophyte nucleomorph: Nature’s smallest nucleus Paul R. Gilson*†, Vanessa Su†‡, Claudio H. Slamovits§, Michael E. Reith¶, Patrick J. Keeling§, and Geoffrey I. McFadden‡ʈ *Infection and Immunity Division, The Walter and Eliza Hall Institute of Medical Research, Parkville 3050, Australia; ‡School of Botany, University of Melbourne, Victoria 3010, Australia; ¶Institute for Marine Biosciences, National Research Council, Halifax, NS, Canada B3H 3Z1; and §Department of Botany, University of British Columbia, Vancouver, BC, Canada V6T 1Z4 Edited by Jeffrey D. Palmer, Indiana University, Bloomington, IN, and approved April 19, 2006 (received for review January 26, 2006) The introduction of plastids into different heterotrophic protists created lineages of algae that diversified explosively, proliferated in marine and freshwater environments, and radically altered the biosphere. The origins of these secondary plastids are usually inferred from the presence of additional plastid membranes. How- ever, two examples provide unique snapshots of secondary- endosymbiosis-in-action, because they retain a vestige of the endosymbiont nucleus known as the nucleomorph. These are chlorarachniophytes and cryptomonads, which acquired their plas- tids from a green and red alga respectively. To allow comparisons between them, we have sequenced the nucleomorph genome from the chlorarachniophyte Bigelowiella natans: at a mere 373,000 bp Fig. 1. Evolution of chlorarachniophytes such as B. natans by sequential endosymbioses. Enslavement of a photosynthetic, cyanobacterium-like pro- and with only 331 genes, the smallest nuclear genome known and karyote (Cb) introduces photosynthesis into a eukaryotic host (Euk 1), whose a model for extreme reduction. The genome is eukaryotic in nature, nucleus (Nu1) acquires at least 1,000 cyanobacterial genes over time. -

Living Organisms Author Their Read-Write Genomes in Evolution
biology Review Living Organisms Author Their Read-Write Genomes in Evolution James A. Shapiro ID Department of Biochemistry and Molecular Biology, University of Chicago GCIS W123B, 979 E. 57th Street, Chicago, IL 60637, USA; [email protected]; Tel.: +1-773-702-1625 Academic Editor: Andrés Moya Received: 23 August 2017; Accepted: 28 November 2017; Published: 6 December 2017 Abstract: Evolutionary variations generating phenotypic adaptations and novel taxa resulted from complex cellular activities altering genome content and expression: (i) Symbiogenetic cell mergers producing the mitochondrion-bearing ancestor of eukaryotes and chloroplast-bearing ancestors of photosynthetic eukaryotes; (ii) interspecific hybridizations and genome doublings generating new species and adaptive radiations of higher plants and animals; and, (iii) interspecific horizontal DNA transfer encoding virtually all of the cellular functions between organisms and their viruses in all domains of life. Consequently, assuming that evolutionary processes occur in isolated genomes of individual species has become an unrealistic abstraction. Adaptive variations also involved natural genetic engineering of mobile DNA elements to rewire regulatory networks. In the most highly evolved organisms, biological complexity scales with “non-coding” DNA content more closely than with protein-coding capacity. Coincidentally, we have learned how so-called “non-coding” RNAs that are rich in repetitive mobile DNA sequences are key regulators of complex phenotypes. Both biotic and abiotic ecological challenges serve as triggers for episodes of elevated genome change. The intersections of cell activities, biosphere interactions, horizontal DNA transfers, and non-random Read-Write genome modifications by natural genetic engineering provide a rich molecular and biological foundation for understanding how ecological disruptions can stimulate productive, often abrupt, evolutionary transformations.