Shared Strategies for Β-Lactam Catabolism in the Soil Microbiome
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Generated by SRI International Pathway Tools Version 25.0, Authors S
Authors: Pallavi Subhraveti Ron Caspi Quang Ong Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000725805Cyc: Streptomyces xanthophaeus Cellular Overview Connections between pathways are omitted for legibility. -

Collection of Information on Enzymes a Great Deal of Additional Information on the European Union Is Available on the Internet
European Commission Collection of information on enzymes A great deal of additional information on the European Union is available on the Internet. It can be accessed through the Europa server (http://europa.eu.int). Luxembourg: Office for Official Publications of the European Communities, 2002 ISBN 92-894-4218-2 © European Communities, 2002 Reproduction is authorised provided the source is acknowledged. Final Report „Collection of Information on Enzymes“ Contract No B4-3040/2000/278245/MAR/E2 in co-operation between the Federal Environment Agency Austria Spittelauer Lände 5, A-1090 Vienna, http://www.ubavie.gv.at and the Inter-University Research Center for Technology, Work and Culture (IFF/IFZ) Schlögelgasse 2, A-8010 Graz, http://www.ifz.tu-graz.ac.at PROJECT TEAM (VIENNA / GRAZ) Werner Aberer c Maria Hahn a Manfred Klade b Uli Seebacher b Armin Spök (Co-ordinator Graz) b Karoline Wallner a Helmut Witzani (Co-ordinator Vienna) a a Austrian Federal Environmental Agency (UBA), Vienna b Inter-University Research Center for Technology, Work, and Culture - IFF/IFZ, Graz c University of Graz, Department of Dermatology, Division of Environmental Dermatology, Graz Executive Summary 5 EXECUTIVE SUMMARY Technical Aspects of Enzymes (Chapter 3) Application of enzymes (Section 3.2) Enzymes are applied in various areas of application, the most important ones are technical use, manufacturing of food and feedstuff, cosmetics, medicinal products and as tools for re- search and development. Enzymatic processes - usually carried out under mild conditions - are often replacing steps in traditional chemical processes which were carried out under harsh industrial environments (temperature, pressures, pH, chemicals). Technical enzymes are applied in detergents, for pulp and paper applications, in textile manufacturing, leather industry, for fuel production and for the production of pharmaceuticals and chiral substances in the chemical industry. -

Download Author Version (PDF)
Food & Function Accepted Manuscript This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. We will replace this Accepted Manuscript with the edited and formatted Advance Article as soon as it is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/foodfunction Page 1 of 16 PleaseFood do not & Functionadjust margins Food&Function REVIEW ARTICLE Interactions between acrylamide, microorganisms, and food components – a review. Received 00th January 20xx, a† a a a a Accepted 00th January 20xx A. Duda-Chodak , Ł. Wajda , T. Tarko , P. Sroka , and P. Satora DOI: 10.1039/x0xx00000x Acrylamide (AA) and its metabolites have been recognised as potential carcinogens, but also they can cause other negative symptoms in human or animal organisms so this chemical compounds still attract a lot of attention. Those substances are www.rsc.org/ usually formed during heating asparagine in the presence of compounds that have α-hydroxycarbonyl groups, α,β,γ,δ- diunsaturated carbonyl groups or α-dicarbonyl groups. -

298009833.Pdf
CORE Metadata, citation and similar papers at core.ac.uk Provided by NOPR Indian Journal of Biotechnology Vol. 18, July 2019, pp 260-268 Mathematically optimized production, purification and characterization of penicillin G acylase from soil bacterial isolates AA17A and AA17B Abhishek Ajamani1 Rajesh Kumar1, Prachi Bhargava2 and Siddharth Vats2* 1University Institute of Engineering and Technology, Kurukshetra University, Kurukshetra 136 119; 2Faculty of Biotechnology, IBST, Shri Ramswaroop Memorial University, Lucknow 225 003 Received 31 January 2019 ; revised 27 March 2019 ; accepted 5 April 2019 This research article deals with production of industrial enzyme penicillin G acylase from soil bacterial isolates namely AA17A and AA17B, which are selected from 80 soil samples. The strains were selected based on qualitative (turbidity) and quantitative (HPLC) test for 6-aminopenicillanic acid (6APA) production. The enzyme was assayed for its activity and optimized for production of enzyme using design of experiments software (DOE) “Design Expert 8.0.7.1”. Optimization of enzyme production of four carbon sources (glucose, glycerol, sucrose and starch), four nitrogen sources (beef extract, tryptone, peptone and yeast extract), for temperature (25°C, 30°C, 35°C and 40°C), four pH (6, 7, 8, 9), four inoculum volumes (2.5 ml, 5.0 ml, 7.5 ml, 10.0 ml) and the phenyl acetic acid (PAA) level (0.15%, 0.17%, 0.185%, 0.2%). The penicillin acylase activity was enhanced to 1.2 fold under following optimized culture conditions: carbon source - glucose (8%), nitrogen source - beef extract (2%), pH 9.0, temperature 30ºC, phenyl acetic acid 0.185%, inoculum volume 5 ml. -

Transcriptome Analysis of Pseudomonas Aeruginosa Biofilm Infection in an Ex Vivo Pig Model of the Cystic Fibrosis Lung
Transcriptome analysis of Pseudomonas aeruginosa biofilm infection in an ex vivo pig model of the cystic fibrosis lung. Niamh E. Harringtona*, Jenny L. Littlerb, Freya Harrisonc* a,b,c: School of Life Sciences, Gibbet Hill Campus, The University of Warwick, Coventry, CV4 7AL, United Kingdom a: [email protected] b: [email protected] c: [email protected] *Co-corresponding authors (Niamh Harrington, Freya Harrison) SUPPLEMENTARY FIGURES AND TABLES Figure S1. A diagram and photograph demonstrating the two environments of the ex vivo pig lung (EVPL) model from which Pseudomonas aeruginosa PA14 RNA was extracted and sequenced: the lung tissue and surrounding synthetic cystic fibrosis sputum media (SCFM). A 400 µl SCFM-agarose pad was made in each well of 24-well plates, and then the EVPL bronchiolar tissue was dissected into ~5 mm x 5 mm squares. The lung tissue pieces were placed on top of the SCFM-agarose pad and infected with PA14, with uninfected tissue used as a control. Each lung piece was then surrounded by 500 µl SCFM (surrounding SCFM) and the plate covered with a Breathe-Easier® membrane (Diversified Biotech), and incubated at 37 °C for the desired length of time. The surrounding SCFM was removed from the well for RNA extraction and the lung tissue piece bead beaten to retrieve the associated biofilm for RNA extraction. Table S1. The concentration of RNA (ng µl-1) extracted from 6 replicate in vitro synthetic cystic fibrosis sputum media (SCFM) Pseudomonas aeruginosa PA14 cultures following 7 d incubation at 37 °C. -

Synthesis, Modification and Characterisation of Magnetic Micro-Matrices for Covalent Immobilisation of Biomolecules. Model Investigations with Penicillin Amidase from E. Coli
Synthesis, modification and characterisation of magnetic micro-matrices for covalent immobilisation of biomolecules. Model investigations with penicillin amidase from E.coli Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften der Naturwissenschaftlichen Fakultät IV – Chemie und Pharmazie der Universität Regensburg vorgelegt von Daniela Petrova Bozhinova aus Plovdiv, Bulgarien 2004 Promotionsgesuch eingereicht am: Die Arbeit wurde angeleitet von: Prof. Dr. habil. Rainer Köster Universität Regensburg, Forschungszentrum Karlsruhe Begutachtung: 1. Gutachter: Prof. Dr. habil Rainer Köster 2. Gutachter: Prof. fil. dr. Volker Kasche Prüfungsausschuss: Vorsitz: Prof. Dr. habil. Hans-Helmut Kohler Universität Regensburg Prüfer: Prof. Dr. habil Rainer Köster Universität Regensburg, Forschungszentrum Karlsruhe Prüfer: Prof. fil. dr. Volker Kasche Technische Universität Hamburg-Harburg Prüfer: Prof. Dr. habil. Claudia Steinem Universität Regensburg To my grandmother Dimitria На баба ми Димитрия ZUSAMMENFASSUNG Aus überwiegend ökonomischen Gründen ist die Immobilisierung und der wiederholte Einsatz von Enzymen in industriellen biotechnologischen Anwendungen erforderlich. In den letzten Jahren wurden magnetische Trägermaterialien, die in vielen Trennverfahren effizient verwendet werden können, in der Biotechnologie, Molekularbiologie und Medizin etabliert. Das Ziel dieser Arbeit war die Entwicklung und Charakterisierung von magnetischen Mikroträgern für die kovalente Immobilisierung von Biokatalysatoren. Als Modellenzym wurde Penicillin -

Streptomyces Coelicolor Strains Lacking Polyprenol Phosphate Mannose Synthase and Protein O-Mannosyl Transferase Are Hyper-Susceptible to Multiple Antibiotics
This is a repository copy of Streptomyces coelicolor strains lacking polyprenol phosphate mannose synthase and protein O-mannosyl transferase are hyper-susceptible to multiple antibiotics. White Rose Research Online URL for this paper: https://eprints.whiterose.ac.uk/128020/ Version: Accepted Version Article: Howlett, Robert, Read, Nicholas, Varghese, Anpu S. et al. (3 more authors) (2018) Streptomyces coelicolor strains lacking polyprenol phosphate mannose synthase and protein O-mannosyl transferase are hyper-susceptible to multiple antibiotics. Microbiology. pp. 369-382. ISSN 1465-2080 https://doi.org/10.1099/mic.0.000605 Reuse Items deposited in White Rose Research Online are protected by copyright, with all rights reserved unless indicated otherwise. They may be downloaded and/or printed for private study, or other acts as permitted by national copyright laws. The publisher or other rights holders may allow further reproduction and re-use of the full text version. This is indicated by the licence information on the White Rose Research Online record for the item. Takedown If you consider content in White Rose Research Online to be in breach of UK law, please notify us by emailing [email protected] including the URL of the record and the reason for the withdrawal request. [email protected] https://eprints.whiterose.ac.uk/ Microb iology Streptomyces coelicolor strains lacking polyprenol phosphate mannose synthase and protein O-mannosyl transferase are hyper-susceptible to multiple antibiotics --Manuscript Draft-- Manuscript -

PRODUCTION, IMMOBILIZATION and INDUSTRIAL USES of PENICILLIN G IJCRR Section: Healthcare ACYLASE Sci
Review Article PRODUCTION, IMMOBILIZATION AND INDUSTRIAL USES OF PENICILLIN G IJCRR Section: Healthcare ACYLASE Sci. Journal Impact Factor 4.016 Mohamed E. Hassan1,2 1Natural and Microbial Products Chemistry Department, National Research Centre, 12622 Dokki, Giza, Egypt; 2Center of Excellence, Advanced Material & Nanobiotechnology Group, National Research Centre, 12622 Dokki, Giza, Egypt. ABSTRACT Penicillin Gacylase in one of the most important enzymes, it belonging to β-lactam antibiotics, first report on the enzyme penicillin acylase was in 1950 when they found in the mycelium of a Penicillium sp. The enzyme appeared to be a periplasmaticheter- odimeric N-terminal serinehydrolase with a molecular mass of 86,183 Da, with a 23,817 Da (209 amino acids)α-subunit and a 62,366 Da (566 amino acids) β-subunit. This enzyme is capable of hydrolyzing penicillin G into phenyl acetic acid and 6-amin- openicillanic acid (6-APA) so this enzyme isthe starting material for the manufacture of penicillin derivatives, which are the most widely used β-lactam antibiotics. Both natural and semi-synthetic penicillins contain 6-aminopenicillanic acid. Key Words: β-lactamantibiotics, Penicillin gacylase, Classification, Industrial uses. INTRODUCTION The basic structure of the penicillins is 6-aminopenicillanic acid (6-APA)(Fig.1), which consists of a thiazolidine ring The β-lactam antibiotics with a condensed β-lactam ring. The 6-APA carries a vari- able acyl moiety (side chain) in position 6. If the penicillin One of the most important groups of antibiotics, both histori- fermentation is carried out without addition of side-chain cally and medically, is the β-lactam group. -
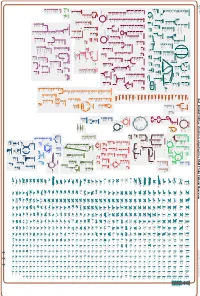
Generate Metabolic Map Poster
Authors: Pallavi Subhraveti Ron Caspi Peter Midford Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_000263195Cyc: Emticicia oligotrophica DSM 17448 Cellular Overview Connections between pathways are omitted for legibility. -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S. -

Targeting Enzymes for Cancer Therapy: Old Enzymes in New Roles
Br. J. Cancer (I994), 70, 786 794 C) Macmifan Press Ltd., 1994 Br.~~ J. Cacr(94,7,7674CMcilnPesLd,19 REVIEW Targeting enzymes for cancer therapy: old enzymes in new roles M.P. Deonarain & A.A. Epenetos Tumour Targeting Laboratory, ICRF Oncology Unit, Royal Postgraduate Medical School, Hammersmith Hospital, Du Cane Road, London W12 OHS, UK. S..y Enzymes which traditionally have played no role in cel-irected cytotoxicity are finding their way into schemes for prodrug activation and immunotoxins owing to such usefl enzymatic activity. Alaline phophatase, carboxypeptidases, D-ghucosiaes and P-lactamases among many others are being utiised to regenrate potent anti-caner drugs or toxic small molecusl from precursors in a bid to enhance their activity in tumours. These prodrug activation systems reqmre the pretargeting of the enzyme to the surface of a tumour cel, usually by an antibody or its immunoreactive fragment. A recent novel approach proposes the intracllular delivery of approprite enzymes, such as phosphodiesterases, to particular celular compartments. Tbere, enzyme activity can cause substantie damag resulting in cell death. Cell targetig of mammahan phosphodiesterases promises to improve upon conventional immunotoxins because of their increased cytotox- icity when tared to the apropte cmpartment and their expected lack of, or lower, immunogenicity in clinical use, The toxicities associated with conventional cancer prodrug activation systems. Such antigens are human car- chemotherapy arise primarily from the lack of specificity for cinoembryonic antigen (CEA) and human chorionic gonado- tumour cells. Most of the presently available drugs are tropin (hCG). designed to be selectively toxic to rapidly dividing cells Monoclonal antibodies remain the choice vehicle for (Valeriote & Putten, 1975). -

(12) United States Patent (10) Patent No.: US 8,561,811 B2 Bluchel Et Al
USOO8561811 B2 (12) United States Patent (10) Patent No.: US 8,561,811 B2 Bluchel et al. (45) Date of Patent: Oct. 22, 2013 (54) SUBSTRATE FOR IMMOBILIZING (56) References Cited FUNCTIONAL SUBSTANCES AND METHOD FOR PREPARING THE SAME U.S. PATENT DOCUMENTS 3,952,053 A 4, 1976 Brown, Jr. et al. (71) Applicants: Christian Gert Bluchel, Singapore 4.415,663 A 1 1/1983 Symon et al. (SG); Yanmei Wang, Singapore (SG) 4,576,928 A 3, 1986 Tani et al. 4.915,839 A 4, 1990 Marinaccio et al. (72) Inventors: Christian Gert Bluchel, Singapore 6,946,527 B2 9, 2005 Lemke et al. (SG); Yanmei Wang, Singapore (SG) FOREIGN PATENT DOCUMENTS (73) Assignee: Temasek Polytechnic, Singapore (SG) CN 101596422 A 12/2009 JP 2253813 A 10, 1990 (*) Notice: Subject to any disclaimer, the term of this JP 2258006 A 10, 1990 patent is extended or adjusted under 35 WO O2O2585 A2 1, 2002 U.S.C. 154(b) by 0 days. OTHER PUBLICATIONS (21) Appl. No.: 13/837,254 Inaternational Search Report for PCT/SG2011/000069 mailing date (22) Filed: Mar 15, 2013 of Apr. 12, 2011. Suen, Shing-Yi, et al. “Comparison of Ligand Density and Protein (65) Prior Publication Data Adsorption on Dye Affinity Membranes Using Difference Spacer Arms'. Separation Science and Technology, 35:1 (2000), pp. 69-87. US 2013/0210111A1 Aug. 15, 2013 Related U.S. Application Data Primary Examiner — Chester Barry (62) Division of application No. 13/580,055, filed as (74) Attorney, Agent, or Firm — Cantor Colburn LLP application No.