Molecular Phylogenetics and Evolution 64 (2012) 255–260
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Literature Cited in Lizards Natural History Database
Literature Cited in Lizards Natural History database Abdala, C. S., A. S. Quinteros, and R. E. Espinoza. 2008. Two new species of Liolaemus (Iguania: Liolaemidae) from the puna of northwestern Argentina. Herpetologica 64:458-471. Abdala, C. S., D. Baldo, R. A. Juárez, and R. E. Espinoza. 2016. The first parthenogenetic pleurodont Iguanian: a new all-female Liolaemus (Squamata: Liolaemidae) from western Argentina. Copeia 104:487-497. Abdala, C. S., J. C. Acosta, M. R. Cabrera, H. J. Villaviciencio, and J. Marinero. 2009. A new Andean Liolaemus of the L. montanus series (Squamata: Iguania: Liolaemidae) from western Argentina. South American Journal of Herpetology 4:91-102. Abdala, C. S., J. L. Acosta, J. C. Acosta, B. B. Alvarez, F. Arias, L. J. Avila, . S. M. Zalba. 2012. Categorización del estado de conservación de las lagartijas y anfisbenas de la República Argentina. Cuadernos de Herpetologia 26 (Suppl. 1):215-248. Abell, A. J. 1999. Male-female spacing patterns in the lizard, Sceloporus virgatus. Amphibia-Reptilia 20:185-194. Abts, M. L. 1987. Environment and variation in life history traits of the Chuckwalla, Sauromalus obesus. Ecological Monographs 57:215-232. Achaval, F., and A. Olmos. 2003. Anfibios y reptiles del Uruguay. Montevideo, Uruguay: Facultad de Ciencias. Achaval, F., and A. Olmos. 2007. Anfibio y reptiles del Uruguay, 3rd edn. Montevideo, Uruguay: Serie Fauna 1. Ackermann, T. 2006. Schreibers Glatkopfleguan Leiocephalus schreibersii. Munich, Germany: Natur und Tier. Ackley, J. W., P. J. Muelleman, R. E. Carter, R. W. Henderson, and R. Powell. 2009. A rapid assessment of herpetofaunal diversity in variously altered habitats on Dominica. -

Male Courtship Display in Two Populations of Anolis Heterodermus (Squamata: Dactyloidae) from the Eastern Cordillera of Colombia
Herpetology Notes, volume 12: 881-884 (2019) (published online on 15 August 2019) Male courtship display in two populations of Anolis heterodermus (Squamata: Dactyloidae) from the Eastern Cordillera of Colombia Iván Beltrán1,2,* and Leidy Alejandra Barragán-Contreras3 Animal displays are generally associated with Anolis heterodermus (Duméril, 1851) is a medium-size territoriality, predator avoidance and courtship arboreal lizard that inhabits shrubs and small trees of high behaviour, in which visual cues transmit a large amount Andean forests in Colombia and Ecuador (Moreno-Arias of information (Alcock and Rubenstein, 1989). Visual and Urbina-Cardona, 2013). Their aggressive and sexual cues can vary in type and frequency depending on behaviour have been described mainly as occasional several factors such as habitat structure, environmental observations in the field and laboratory (Jenssen, 1975; temperature and density of conspecifics (Endler, 1992; Guzmán, 1989; Beltrán, 2019). This species belongs to Candolin, 2003). Visual displays usually convey the heterodermus complex of species from which its information about species identity and/or physiological phylogenetic relations are not well established (Lazell, status of the signaller. Moreover, since an effective 1969; Castañeda and de Queiroz, 2013). Recently, it communication will determine the reproductive was suggested that there are at least three genetically success of the individual and ultimately its fitness, the distinct clades within the complex (Vargas-Ramírez and information must be quickly comprehended by the Moreno-Arias, 2014). However, there is no evidence receiver (Sullivan and Kwiatkowski, 2007). Variations that these genetic differences are backed by behavioural in the signalling pathway constitute a prezygotic changes that could act as a prezygotic barrier. -

The Evolution of Androgen Receptor Expression and Behavior in Anolis Lizard Forelimb Muscles
Trinity University Digital Commons @ Trinity Biology Faculty Research Biology Department 2018 The Evolution of Androgen Receptor Expression and Behavior in Anolis Lizard Forelimb Muscles Michele A. Johnson Trinity University, [email protected] Bonnie K. Kircher [email protected] Diego J. Castro Trinity University Follow this and additional works at: https://digitalcommons.trinity.edu/bio_faculty Part of the Biology Commons Repository Citation Johnson, M.A., Kircher, B.K., & Castro, D.J. (2018). The evolution of androgen receptor expression and behavior in Anolis lizard forelimb muscles. Journal of Comparative Physiology A: Neuroethology, Sensory, Neural, and Behavioral Physiology, 204(1), 71-79. doi:10.1007/s00359-017-1228-y This Article is brought to you for free and open access by the Biology Department at Digital Commons @ Trinity. It has been accepted for inclusion in Biology Faculty Research by an authorized administrator of Digital Commons @ Trinity. For more information, please contact [email protected]. Author's personal copy Journal of Comparative Physiology A (2018) 204:71–79 https://doi.org/10.1007/s00359-017-1228-y ORIGINAL PAPER The evolution of androgen receptor expression and behavior in Anolis lizard forelimb muscles Michele A. Johnson1 · Bonnie K. Kircher2 · Diego J. Castro1,3 Received: 1 June 2017 / Revised: 12 October 2017 / Accepted: 9 November 2017 / Published online: 15 November 2017 © Springer-Verlag GmbH Germany, part of Springer Nature 2017 Abstract The motor systems that produce behavioral movements are among the primary targets for the action of steroid hormones, including androgens. Androgens such as testosterone bind to androgen receptors (AR) to induce physiological changes in the size, strength, and energetic capacity of skeletal muscles, which can directly influence the performance of behaviors in which those muscles are used. -

Anolis Newsletter VI
Anolis Newsletter VI Edited by D. Luke Mahler Anthony Herrel Jonathan B. Losos i June 2, 2010 The Museum of Comparative Zoology Harvard University 26 Oxford St. Cambridge, MA 02138 USA All rights reserved. Names or nomenclatural acts in this work are disclaimed for nomenclatural purposes under ICZN 8.3. Front cover: The enigmatic, rostrally-endowed Anolis proboscis, from Ecuador. Reprinted with permission from Williams (1979; Breviora 449:1-19). Illustration by Laszlo Meszoly. ii In Memory of A. Stanley Rand (1932-2005) Stan Rand (left) with his former graduate advisor, Ernest Williams (right) at Soroa, Cuba in 1983. iii Preface On the first weekend of October in 2009, 125 anole biologists traveled from eight countries to Harvard University’s Museum of Comparative Zoology to attend the 6th Anolis Symposium. It had been 10 years since the previous symposium, and a reunion was long past due. In 2008, as we began to consider how to proceed with such an endeavor, a fortunate thing happened: the Herpetology Department at the MCZ renovated its library and teaching space – the famous lair of the late pater anolis, Ernest Williams. The library needed a namesake, and Ernest was under strong consideration (after all, he had been instrumental in filling its shelves!). After a brief period of friendly deliberation, it was decided that the library would be dedicated to Williams, and that the occasion would be the commencement of the 6th Anolis Symposium, held at the Museum of Comparative Zoology. Anole biology has changed considerably in the last decade, and it’s been for the better! First and foremost, the field has grown explosively. -

ARE DEWLAP COLOR and DISPLAY BEHAVIOR HONEST INDICATORS of MALE QUALITY in ANOLIS LIZARDS? Ellee G
Trinity University Digital Commons @ Trinity Biology Honors Theses Biology Department 4-19-2013 ARE DEWLAP COLOR AND DISPLAY BEHAVIOR HONEST INDICATORS OF MALE QUALITY IN ANOLIS LIZARDS? Ellee G. Cook Trinity University, [email protected] Follow this and additional works at: http://digitalcommons.trinity.edu/bio_honors Recommended Citation Cook, Ellee G., "ARE DEWLAP COLOR AND DISPLAY BEHAVIOR HONEST INDICATORS OF MALE QUALITY IN ANOLIS LIZARDS?" (2013). Biology Honors Theses. 13. http://digitalcommons.trinity.edu/bio_honors/13 This Thesis open access is brought to you for free and open access by the Biology Department at Digital Commons @ Trinity. It has been accepted for inclusion in Biology Honors Theses by an authorized administrator of Digital Commons @ Trinity. For more information, please contact [email protected]. ARE DEWLAP COLOR AND DISPLAY BEHAVIOR HONEST INDICATORS OF MALE QUALITY IN ANOLIS LIZARDS? ELLEE G. COOK A DEPARTMENT HONORS THESIS SUBMITTED TO THE DEPARTMENT OF BIOLOGY AT TRINITY UNIVERSITY IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR GRADUATION WITH DEPARTMENTAL HONORS DATE ______ ____________________________ ________________________________ THESIS ADVISOR THESIS CO-ADVISOR ________________________________ DEPARTMENT CHAIR _________________________________________________ ASSOCIATE VICE PRESIDENT FOR ACADEMIC AFFAIRS, CURRICULUM AND STUDENT ISSUES Student Copyright Declaration: the author has selected the following copyright provision (select only one): [X] This thesis is licensed under the Creative Commons Attribution-NonCommercial-NoDerivs License, which allows some noncommercial copying and distribution of the thesis, given proper attribution. To view a copy of this license, visit http://creativecommons.org/licenses/ or send a letter to Creative Commons, 559 Nathan Abbott Way, Stanford, California 94305, USA. [ ] This thesis is protected under the provisions of U.S. -

Anolis Tandai, a New Dietary Record for the Amazon Ringed Snake, Rhinobothryum Lentiginosum (Scopoli, 1785) (Squamata: Colubridae)
Herpetology Notes, volume 13: 981-987 (2020) (published online on 26 November 2020) Anolis tandai, a new dietary record for the Amazon ringed snake, Rhinobothryum lentiginosum (Scopoli, 1785) (Squamata: Colubridae) Luis A. García–Ayachi1,2,3,*, Mallory Wittwer3, and Christopher Kirkby3 The Amazon banded snake is a rare, infrequently because they observed these snakes on the ground observed species of squamate with only a few (Martins and Oliveira, 1998; Doan and Arriaga, 2000; specimens stored in biological collections, despite its Duellman, 2005; Ribeiro Duarte, 2010; Avila–Pires et widespread distribution (Cunha and Nascimento, 1978). al., 2010). However, dietary information suggests that R. This snake lives in the lowland Amazonian forests of lentiginosum is a lizard specialist that feeds on a variety Brazil, Venezuela, Guyana, Suriname, French Guiana, of terrestrial and arboreal species that range in size from Colombia, Ecuador, Paraguay, Bolivia, and Peru (Peters small species, such as Gonatodes spp. to the larger and Orejas–Miranda, 1970; Miranda et al., 2009; Iguana iguana (Table 1). Rhinobothryum lentiginosum Reynolds and MacCulloch, 2012; Uetz et al., 2020). likely forages in tree and bushes, and possibly preys Although it has been recorded in primary forest (Cunha on night–sleeping lizards (Oliveira and Martins, 1998; and Nascimento, 1978; Cunha et al., 1985; Martins Martins and Oliveira, 1998; Vitt et al., 2000; Duellman, and Oliveira, 1998; Fraga et al., 2014), some records 2005; Ribeiro Duarte, 2010), and occasionally on frogs, demonstrate that R. lentiginosum also inhabits secondary small birds and mammals (Zimmerman and Rodrigues, forest (Venegas and Couceiro, 2017), and more open, 1990; Avila–Pires et al., 2010). -

Morfología, Distribución Geográfica Y Microhábitat De Los Lagartos Cubanos Del Género Anolis (Lepidosauria: Iguania)
INSTITUTO DE ECOLOGÍA Y SISTEMÁTICA MINISTERIO DE CIENCIA, TECNOLOGÍA Y MEDIO AMBIENTE Morfología, distribución geográfica y microhábitat de los lagartos cubanos del género Anolis (Lepidosauria: Iguania) AUTORA: Lic. Lourdes Rodríguez Schettino TUTOR: Dr. Alberto Coy Otero Tesis en opción al grado científico de Doctor en Ciencias Biológicas La Habana 1999 A mis padres, esposo e hijos SÍNTESIS En el género Anolis Daudin, 1802, se incluyen más de 250 especies, distribuidas desde Norte hasta Sur América, incluyendo las Antillas Mayores y Menores, las Islas Bahamas y algunas islas del Pacífico. En Cuba viven 51 especies (94.1 % endémicas), número que es el mayor registrado para un país. Para conservarlas adecuadamente, los objetivos trazados fueron: preparar una clave para su identificación y verificar la presencia de grupos de especies morfológicamente afines; actualizar la distribución geográfica y altitudinal; caracterizar los tipos de substratos que utilizan y comprobar si existe distribución vertical sobre ellos. Para alcanzarlos, se revisó la literatura al respecto, se tomaron diferentes medidas a los ejemplares depositados en colecciones, se revisaron los ficheros de las colecciones, se visitaron numerosas localidades a través de todo el país, donde se realizaron censos de las especies y observaciones sobre el tipo de substrato y la altura sobre el suelo a que se encontraban. Con todo este trabajo se logró por primera vez: preparar un compendio sobre las especies cubanas del género; identificarlas mediante la clave confeccionada; comprobar -

Hot Trade in Cool Creatures
HOT TRADE IN COOL CREATURES A review of the live reptile trade in the European Union in the 1990s with a focus on Germany by MARK AULIYA A TRAFFIC EUROPE REPORT This report was published with the kind support of Published by TRAFFIC Europe, Brussels, Belgium. © 2003 TRAFFIC Europe All rights reserved. All material appearing in this publication is copyrighted and may be produced with permission. Any reproduction in full or in part of this publication must credit TRAFFIC Europe as the copyright owner. The views of the author expressed in this publication do not necessarily reflect those of the TRAFFIC network, WWF or IUCN. The designations of geographical entities in this publication, and the presentation of the material, do not imply the expression of any opinion whatsoever on the part of TRAFFIC or its supporting organizations concerning the legal status of any country, territory, or area, or of its authorities, or concerning the delimitation of its frontiers or boundaries. The TRAFFIC symbol copyright and Registered Trademark ownership is held by WWF. TRAFFIC is a joint programme of WWF and IUCN. Suggested citation: Auliya, Mark. (2003). Hot trade in cool creatures: A review of the live reptile trade in the European Union in the 1990s with a focus on Germany. TRAFFIC Europe, Brussels, Belgium ISBN 2 9600505 9 2 EAN code: 9782960050592 Front cover photograph: The Green-eyed Gecko Gekko smithii from southern Sumatra. Photograph credit: Mark Auliya Printed on recycled paper HOT TRADE IN COOL CREATURES A REVIEW OF THE LIVE REPTILE TRADE IN THE EUROPEAN UNION IN THE 1990s WITH A FOCUS ON GERMANY The Yellow Monitor Varanus melinus. -

Definition of the Caribbean Islands Biogeographic Region, with Checklist and Recommendations for Standardized Common Names of Amphibians and Reptiles
caribbean herpetology article Definition of the Caribbean Islands biogeographic region, with checklist and recommendations for standardized common names of amphibians and reptiles S. Blair Hedges1,*, Robert Powell2, Robert W. Henderson3, Sarah Hanson1, and John C. Murphy4 1Center for Biodiversity, Temple University, 1925 N. 12th Street, Philadelphia, Pensylvania 19122, USA. 2Department of Biology, Avila University, 11901 Wornall Road, Kansas City, Missouri 64145, USA. 3Vertebrate Zoology, Milwaukee Public Museum, 800 West Wells Street, Milwaukee, Wisconsin 53233, USA. 4Science and Education, Field Museum of Natural History, 1400 Lake Shore Drive, Chicago, IL 60616 USA. *Corresponding author ([email protected]) Edited by: R. Graham Reynolds. Date of publication: 28 May 2019. Citation: Hedges SB, Powell R, Henderson RW, Hanson S, and Murphy JC. 2019. Definition of the Caribbean Islands biogeographic region, with checklist and recommendations for standardized common names of amphibians and reptiles. Caribbean Herpetology, 67, 1–53. DOI: 10.31611/ch.67 Abstract To facilitate biological study we define “Caribbean Islands” as a biogeographic region that includes the An- tilles, the Bahamas, and islands bordering Central and South America separated from mainland areas by at least 20 meters of water depth. The advantages of this definition are that it captures nearly all islands with endemic species and with at least some Antillean-derived species, and still circumscribes a region of high biodiversity and biogeographic significance. We argue that Caribbean islands, in this expanded sense, are also cohesive from a conservation standpoint in that they share high human population densities and sim- ilar conservation threats. A disadvantage of this definition, strictly applied, is that it includes some islands (e.g., Trinidad) that have mostly mainland species. -

The Evolution and Diversity of the Anolis Dewlap
The evolution and diversity of the Anolis dewlap The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Harrison, Alexis Stephania. 2014. The evolution and diversity of the Anolis dewlap. Doctoral dissertation, Harvard University. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:13068393 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA The evolution and diversity of the Anolis dewlap A dissertation presented by Alexis Stephania Harrison to The Department of Organismic and Evolutionary Biology In partial fulfillment of the requirements for the degree of Doctor of Philosophy in the subject of Biology Harvard University Cambridge, Massachusetts September 2014 Alexis Stephania Harrison All rights reserved. ii Thesis Advisor Author Jonathan B. Losos Alexis Stephania Harrison The evolution and diversity of the Anolis dewlap Abstract The neotropical lizard genus Anolis is an important model system for studies of the ecology and evolution of animal diversity. One of the most striking elements of Anolis diversity is found in the morphology of the dewlap, an extensible flap of colored skin on the throat that anoles use to communicate during social interactions. The evolutionary forces that have promoted the evolution of dewlap diversity are poorly understood. A study of reproductive success in A. carolinensis showed for the first time that dewlap color is currently under selection in an anole (Chapter 1). -
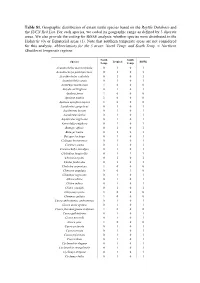
Table S1. Geographic Distribution of Extant Turtle Species Based on the Reptile Database and the IUCN Red List
Table S1. Geographic distribution of extant turtle species based on the Reptile Database and the IUCN Red List. For each species, we coded its geographic range as defined by 3 discrete areas. We also provide the coding for BiSSE analysis: whether species were distributed in the Holarctic (0) or Equatorial areas (1). Note that southern temperate areas are not considered for this analysis. Abbreviations for the 3 areas: North Temp. and South Temp. = Northern (Southern) temperate regions. North South Species Tropical BiSSE Temp. Temp. Acanthochelys macrocephala 0 1 0 1 Acanthochelys pallidipectoris 0 1 0 1 Acanthochelys radiolata 0 1 0 1 Acanthochelys spixii 0 1 0 1 Actinemys marmorata 1 0 0 0 Amyda cartilaginea 0 1 0 1 Apalone ferox 1 0 0 0 Apalone mutica 1 0 0 0 Apalone spinifera aspera 1 0 0 0 Aspideretes gangeticus 0 1 0 1 Aspideretes hurum 0 1 0 1 Aspideretes leithii 0 1 0 1 Aspideretes nigricans 0 1 0 1 Astrochelys yniphora 0 1 0 1 Batagur affinis 0 1 0 1 Batagur baska 0 1 0 1 Batagur kachuga 0 1 0 1 Callagur borneoensis 0 1 0 1 Caretta caretta 0 1 0 1 Carettochelys insculpta 0 1 0 1 Chelodina longicollis 0 1 1 1 Chelonia mydas 0 1 0 1 Chelus fimbriatus 0 1 0 1 Chelydra serpentina 1 0 0 0 Chersine angulata 0 0 1 0 Chinemys nigricans 0 1 0 1 Chitra chitra 0 1 0 1 Chitra indica 0 1 0 1 Chitra vandijki 0 1 0 1 Chrysemys picta 1 0 0 0 Clemmys guttata 1 0 0 0 Cuora amboinensis amboinensis 0 1 0 1 Cuora aurocapitata 0 1 0 1 Cuora flavomarginata evelynae 1 0 0 0 Cuora galbinifrons 0 1 0 1 Cuora mccordi 0 1 0 1 Cuora pani 1 0 0 0 Cuora picturata 0 1 0 1 Cuora serrata 0 1 0 1 Cuora trifasciata 0 1 0 1 Cuora zhoui 0 1 0 1 Cyclanorbis elegans 0 1 0 1 Cyclanorbis senegalensis 0 1 0 1 Cyclemys atripons 0 1 0 1 Cyclemys bellii 0 1 0 1 Cyclemys dentata 0 1 0 1 Cyclemys oldhamii 0 1 0 1 Cyclemys orbiculata 0 1 0 1 Cyclemys ovata 0 1 0 1 Cyclemys pulchristriata 0 1 0 1 Cyclemys shanensis 0 1 0 1 Cyclemys sp. -

Understanding Patterns of Diversity and Evolution in Mainland Anolis Lizards Levi Gray University of New Mexico
University of New Mexico UNM Digital Repository Biology ETDs Electronic Theses and Dissertations Spring 5-11-2018 Understanding patterns of diversity and evolution in mainland Anolis lizards Levi Gray University of New Mexico Follow this and additional works at: https://digitalrepository.unm.edu/biol_etds Part of the Biodiversity Commons, Biology Commons, and the Evolution Commons Recommended Citation Gray, Levi. "Understanding patterns of diversity and evolution in mainland Anolis lizards." (2018). https://digitalrepository.unm.edu/ biol_etds/278 This Dissertation is brought to you for free and open access by the Electronic Theses and Dissertations at UNM Digital Repository. It has been accepted for inclusion in Biology ETDs by an authorized administrator of UNM Digital Repository. For more information, please contact [email protected]. Levi Nathaniel Gray Candidate Department of Biology Department This dissertation is approved, and it is acceptable in quality and form for publication: Approved by the Dissertation Committee: Dr. Steve Poe , Chairperson Dr. Mike Andersen Dr. Joe Cook Dr. Ian Wang i Understanding patterns of diversity and evolution in mainland Anolis lizards By Levi Nathaniel Gray B.S., Evolution, Ecology, and Biodiversity, University of California at Davis, 2006 DISSERTATION Doctor of Philosophy Biology The University of New Mexico Albuquerque, New Mexico July, 2018 ii ACKNOWLEDGEMENTS It is important to me that I recognize the people that have contributed positively towards helping me along the path that led to my degree. This will be tough and I will forget and/or leave some out. Please don’t hold this against me, as I’m writing this with little time left and there is still plenty to finish in my final chapter.