Diversity of Rare and Abundant Prokaryotic
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Microbial Community Structure Dynamics in Ohio River Sediments During Reductive Dechlorination of Pcbs
University of Kentucky UKnowledge University of Kentucky Doctoral Dissertations Graduate School 2008 MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS Andres Enrique Nunez University of Kentucky Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation Nunez, Andres Enrique, "MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS" (2008). University of Kentucky Doctoral Dissertations. 679. https://uknowledge.uky.edu/gradschool_diss/679 This Dissertation is brought to you for free and open access by the Graduate School at UKnowledge. It has been accepted for inclusion in University of Kentucky Doctoral Dissertations by an authorized administrator of UKnowledge. For more information, please contact [email protected]. ABSTRACT OF DISSERTATION Andres Enrique Nunez The Graduate School University of Kentucky 2008 MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS ABSTRACT OF DISSERTATION A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the College of Agriculture at the University of Kentucky By Andres Enrique Nunez Director: Dr. Elisa M. D’Angelo Lexington, KY 2008 Copyright © Andres Enrique Nunez 2008 ABSTRACT OF DISSERTATION MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS The entire stretch of the Ohio River is under fish consumption advisories due to contamination with polychlorinated biphenyls (PCBs). In this study, natural attenuation and biostimulation of PCBs and microbial communities responsible for PCB transformations were investigated in Ohio River sediments. Natural attenuation of PCBs was negligible in sediments, which was likely attributed to low temperature conditions during most of the year, as well as low amounts of available nitrogen, phosphorus, and organic carbon. -

Online Supplementary Figures of Chapter 3
Online Supplementary Figures of Chapter 3 Fabio Gori Figures 1-30 contain pie charts showing the population characterization re- sulting from the taxonomic assignment computed by the methods. On the simulated datasets the true population distribution is also shown. 1 MTR Bacillales (47.11%) Thermoanaerobacterales (0.76%) Clostridiales (33.58%) Lactobacillales (7.99%) Others (10.55%) LCA Bacillales (48.38%) Thermoanaerobacterales (0.57%) Clostridiales (32.14%) Lactobacillales (10.07%) Others (8.84%) True Distribution 333 386 Prochlorales (5.84%) 535 Bacillales (34.61%) Halanaerobiales (4.37%) Thermoanaerobacterales (10.29%) Clostridiales (28.75%) Lactobacillales (9.38%) 1974 Herpetosiphonales (6.77%) 1640 249 587 Figure 1: Population distributions (rank Order) of M1, coverage 0.1x, by MTR and LCA, and the true population distribution. 2 MTR Bacillus (47.34%) Clostridium (14.61%) Lactobacillus (8.71%) Anaerocellum (11.41%) Alkaliphilus (5.14%) Others (12.79%) LCA Bacillus (51.41%) Clostridium (8.08%) Lactobacillus (9.23%) Anaerocellum (15.79%) Alkaliphilus (5.17%) Others (10.31%) True Distribution 386 552 Herpetosiphon (6.77%) 333 Prochlorococcus (5.84%) 587 Bacillus (34.61%) Clostridium (19.07%) Lactobacillus (9.38%) 249 Halothermothrix (4.37%) Caldicellulosiruptor (10.29%) Alkaliphilus (9.68%) 535 1974 1088 Figure 2: Population distributions (rank Genus) of M1, coverage 0.1x, by MTR and LCA, and the true population distribution. 3 MTR Prochlorales (0.07%) Bacillales (47.97%) Thermoanaerobacterales (0.66%) Clostridiales (32.18%) Lactobacillales (7.76%) Others (11.35%) LCA Prochlorales (0.10%) Bacillales (49.02%) Thermoanaerobacterales (0.59%) Clostridiales (30.62%) Lactobacillales (9.50%) Others (10.16%) True Distribution 3293 3950 Prochlorales (5.65%) 5263 Bacillales (36.68%) Halanaerobiales (3.98%) Thermoanaerobacterales (10.56%) Clostridiales (27.34%) Lactobacillales (9.03%) 21382 Herpetosiphonales (6.78%) 15936 2320 6154 Figure 3: Population distributions (rank Order) of M1, coverage 1x, by MTR and LCA, and the true population distribution. -

Metagenomic and PCR-Based Diversity Surveys of [Fefe
ORIGINAL RESEARCH published: 30 August 2016 doi: 10.3389/fmicb.2016.01301 Metagenomic and PCR-Based Diversity Surveys of [FeFe]-Hydrogenases Combined with Isolation of Alkaliphilic Hydrogen-Producing Bacteria from the Serpentinite-Hosted Prony Hydrothermal Field, New Caledonia Nan Mei 1, Anne Postec 1, Christophe Monnin 2, Bernard Pelletier 3, Claude E. Payri 3, Bénédicte Ménez 4, Eléonore Frouin 1, Bernard Ollivier 1, Gaël Erauso 1 and Marianne Quéméneur 1* 1 2 Edited by: Aix Marseille Univ, Université de Toulon, CNRS, IRD, MIO, Marseille, France, GET UMR5563 (Centre National de la 3 Andreas Teske, Recherche Scientifique/UPS/IRD/CNES), Géosciences Environnement Toulouse, Toulouse, France, Institut pour la 4 University of North Carolina at Chapel Recherche et le Développement (IRD) Centre de Nouméa, MIO UM 110, Nouméa, Nouvelle-Calédonie, Institut de Physique Hill, USA du Globe de Paris, Sorbonne Paris Cité, Univ Paris Diderot, Centre National de la Recherche Scientifique, Paris, France Reviewed by: John R. Spear, High amounts of hydrogen are emitted in the serpentinite-hosted hydrothermal field Colorado School of Mines, USA of the Prony Bay (PHF, New Caledonia), where high-pH (∼11), low-temperature Igor Tiago, ◦ University of Coimbra, Portugal (<40 C), and low-salinity fluids are discharged in both intertidal and shallow submarine *Correspondence: environments. In this study, we investigated the diversity and distribution of potentially Marianne Quéméneur hydrogen-producing bacteria in Prony hyperalkaline springs by using metagenomic [email protected] analyses and different PCR-amplified DNA sequencing methods. The retrieved Specialty section: sequences of hydA genes, encoding the catalytic subunit of [FeFe]-hydrogenases and, This article was submitted to used as a molecular marker of hydrogen-producing bacteria, were mainly related to those Extreme Microbiology, a section of the journal of Firmicutes and clustered into two distinct groups depending on sampling locations. -

Global Metagenomic Survey Reveals a New Bacterial Candidate Phylum in Geothermal Springs
ARTICLE Received 13 Aug 2015 | Accepted 7 Dec 2015 | Published 27 Jan 2016 DOI: 10.1038/ncomms10476 OPEN Global metagenomic survey reveals a new bacterial candidate phylum in geothermal springs Emiley A. Eloe-Fadrosh1, David Paez-Espino1, Jessica Jarett1, Peter F. Dunfield2, Brian P. Hedlund3, Anne E. Dekas4, Stephen E. Grasby5, Allyson L. Brady6, Hailiang Dong7, Brandon R. Briggs8, Wen-Jun Li9, Danielle Goudeau1, Rex Malmstrom1, Amrita Pati1, Jennifer Pett-Ridge4, Edward M. Rubin1,10, Tanja Woyke1, Nikos C. Kyrpides1 & Natalia N. Ivanova1 Analysis of the increasing wealth of metagenomic data collected from diverse environments can lead to the discovery of novel branches on the tree of life. Here we analyse 5.2 Tb of metagenomic data collected globally to discover a novel bacterial phylum (‘Candidatus Kryptonia’) found exclusively in high-temperature pH-neutral geothermal springs. This lineage had remained hidden as a taxonomic ‘blind spot’ because of mismatches in the primers commonly used for ribosomal gene surveys. Genome reconstruction from metagenomic data combined with single-cell genomics results in several high-quality genomes representing four genera from the new phylum. Metabolic reconstruction indicates a heterotrophic lifestyle with conspicuous nutritional deficiencies, suggesting the need for metabolic complementarity with other microbes. Co-occurrence patterns identifies a number of putative partners, including an uncultured Armatimonadetes lineage. The discovery of Kryptonia within previously studied geothermal springs underscores the importance of globally sampled metagenomic data in detection of microbial novelty, and highlights the extraordinary diversity of microbial life still awaiting discovery. 1 Department of Energy Joint Genome Institute, Walnut Creek, California 94598, USA. 2 Department of Biological Sciences, University of Calgary, Calgary, Alberta T2N 1N4, Canada. -

Hydrogenophaga Electricum Sp. Nov., Isolated from Anodic Biofilms of an Acetate-Fed Microbial Fuel Cell
J. Gen. Appl. Microbiol., 59, 261‒266 (2013) Full Paper Hydrogenophaga electricum sp. nov., isolated from anodic biofilms of an acetate-fed microbial fuel cell Zen-ichiro Kimura and Satoshi Okabe* Division of Environmental Engineering, Faculty of Engineering, Hokkaido University, Kita-ku, Sapporo, Hokkaido 060‒8628, Japan (Received October 25, 2012; Accepted April 2, 2013) A Gram-negative, non-spore-forming, rod-shaped bacterial strain, AR20T, was isolated from an- odic biofilms of an acetate-fed microbial fuel cell in Japan and subjected to a polyphasic taxo- nomic study. Strain AR20T grew optimally at pH 7.0‒8.0 and 25°C. It contained Q-8 as the pre- dominant ubiquinone and C16:0, summed feature 3 (C16:1ω7c and/or iso-C15:02OH), and C18:1ω7c as the major fatty acids. The DNA G+C content was 67.1 mol%. A neighbor-joining phylogenetic tree revealed that strain AR20T clustered with three type strains of the genus Hydrogenophaga (H. flava, H. bisanensis and H. pseudoflava). Strain AR20T exhibited 16S rRNA gene sequence similarity values of 95.8‒97.7% to the type strains of the genus Hydrogenophaga. On the basis of phenotypic, chemotaxonomic and phylogenetic data, strain AR20T is considered a novel species of the genus Hydrogenophaga, for which the name Hydrogenophaga electricum sp. nov. is pro- posed. The type strain is AR20T (= KCTC 32195T = NBRC 109341T). Key Words—Hydrogenophaga electricum; hydrogenotrophic exoelectrogen; microbial fuel cell Introduction the MFC was analyzed. Results showed that bacteria belonging to the genera Geobacter and Hydrogenoph- Microbial fuel cells (MFCs) are devices that are able aga were abundantly present in the anodic biofilm to directly convert the chemical energy of organic community (Kimura and Okabe, 2013). -

Ample Arsenite Bio-Oxidation Activity in Bangladesh Drinking Water Wells: a Bonanza for Bioremediation?
microorganisms Article Ample Arsenite Bio-Oxidation Activity in Bangladesh Drinking Water Wells: A Bonanza for Bioremediation? 1,2 3 3 1 1, Zahid Hassan , Munawar Sultana , Sirajul I. Khan , Martin Braster , Wilfred F.M. Röling y and Hans V. Westerhoff 1,4,5,* 1 Department of Molecular Cell Biology, Faculty of Science, Vrije Universiteit Amsterdam, 1081 HV Amsterdam, The Netherlands 2 Department of Genetic Engineering and Biotechnology, Jagannath University, Dhaka 1100, Bangladesh 3 Department of Microbiology, University of Dhaka, Dhaka 1000, Bangladesh 4 Manchester Centre for Integrative Systems Biology (MCISB), School of Chemical Engineering and Analytical Sciences (SCEAS), the University of Manchester, Manchester M13 9PL, UK 5 Synthetic Systems Biology and Nuclear Organization, Swammerdam Institute for Life Sciences, University of Amsterdam, 1098 XH Amsterdam, The Netherlands * Correspondence: h.v.westerhoff@vu.nl; Tel.: +31-205-987-230 Deceased 25 September 2015. y Received: 18 June 2019; Accepted: 31 July 2019; Published: 8 August 2019 Abstract: Millions of people worldwide are at risk of arsenic poisoning from their drinking water. In Bangladesh the problem extends to rural drinking water wells, where non-biological solutions are not feasible. In serial enrichment cultures of water from various Bangladesh drinking water wells, we found transfer-persistent arsenite oxidation activity under four conditions (aerobic/anaerobic; heterotrophic/autotrophic). This suggests that biological decontamination may help ameliorate the problem. The enriched microbial communities were phylogenetically at least as diverse as the unenriched communities: they contained a bonanza of 16S rRNA gene sequences. These related to Hydrogenophaga, Acinetobacter, Dechloromonas, Comamonas, and Rhizobium/Agrobacterium species. In addition, the enriched microbiomes contained genes highly similar to the arsenite oxidase (aioA) gene of chemolithoautotrophic (e.g., Paracoccus sp. -

Rapport Nederlands
Moleculaire detectie van bacteriën in dekaarde Dr. J.J.P. Baars & dr. G. Straatsma Plant Research International B.V., Wageningen December 2007 Rapport nummer 2007-10 © 2007 Wageningen, Plant Research International B.V. Alle rechten voorbehouden. Niets uit deze uitgave mag worden verveelvoudigd, opgeslagen in een geautomatiseerd gegevensbestand, of openbaar gemaakt, in enige vorm of op enige wijze, hetzij elektronisch, mechanisch, door fotokopieën, opnamen of enige andere manier zonder voorafgaande schriftelijke toestemming van Plant Research International B.V. Exemplaren van dit rapport kunnen bij de (eerste) auteur worden besteld. Bij toezending wordt een factuur toegevoegd; de kosten (incl. verzend- en administratiekosten) bedragen € 50 per exemplaar. Plant Research International B.V. Adres : Droevendaalsesteeg 1, Wageningen : Postbus 16, 6700 AA Wageningen Tel. : 0317 - 47 70 00 Fax : 0317 - 41 80 94 E-mail : [email protected] Internet : www.pri.wur.nl Inhoudsopgave pagina 1. Samenvatting 1 2. Inleiding 3 3. Methodiek 8 Algemene werkwijze 8 Bestudeerde monsters 8 Monsters uit praktijkteelten 8 Monsters uit proefteelten 9 Alternatieve analyse m.b.v. DGGE 10 Vaststellen van verschillen tussen de bacterie-gemeenschappen op myceliumstrengen en in de omringende dekaarde. 11 4. Resultaten 13 Monsters uit praktijkteelten 13 Monsters uit proefteelten 16 Alternatieve analyse m.b.v. DGGE 23 Vaststellen van verschillen tussen de bacterie-gemeenschappen op myceliumstrengen en in de omringende dekaarde. 25 5. Discussie 28 6. Conclusies 33 7. Suggesties voor verder onderzoek 35 8. Gebruikte literatuur. 37 Bijlage I. Bacteriesoorten geïsoleerd uit dekaarde en van mycelium uit commerciële teelten I-1 Bijlage II. Bacteriesoorten geïsoleerd uit dekaarde en van mycelium uit experimentele teelten II-1 1 1. -

EXPERIMENTAL STUDIES on FERMENTATIVE FIRMICUTES from ANOXIC ENVIRONMENTS: ISOLATION, EVOLUTION, and THEIR GEOCHEMICAL IMPACTS By
EXPERIMENTAL STUDIES ON FERMENTATIVE FIRMICUTES FROM ANOXIC ENVIRONMENTS: ISOLATION, EVOLUTION, AND THEIR GEOCHEMICAL IMPACTS By JESSICA KEE EUN CHOI A dissertation submitted to the School of Graduate Studies Rutgers, The State University of New Jersey In partial fulfillment of the requirements For the degree of Doctor of Philosophy Graduate Program in Microbial Biology Written under the direction of Nathan Yee And approved by _______________________________________________________ _______________________________________________________ _______________________________________________________ _______________________________________________________ New Brunswick, New Jersey October 2017 ABSTRACT OF THE DISSERTATION Experimental studies on fermentative Firmicutes from anoxic environments: isolation, evolution and their geochemical impacts by JESSICA KEE EUN CHOI Dissertation director: Nathan Yee Fermentative microorganisms from the bacterial phylum Firmicutes are quite ubiquitous in subsurface environments and play an important biogeochemical role. For instance, fermenters have the ability to take complex molecules and break them into simpler compounds that serve as growth substrates for other organisms. The research presented here focuses on two groups of fermentative Firmicutes, one from the genus Clostridium and the other from the class Negativicutes. Clostridium species are well-known fermenters. Laboratory studies done so far have also displayed the capability to reduce Fe(III), yet the mechanism of this activity has not been investigated -

Biochemical Characterization of Β-Xylan Acting Glycoside
BIOCHEMICAL CHARACTERIZATION OF β-XYLAN ACTING GLYCOSIDE HYDROLASES FROM THE THERMOPHILIC BACTERIUM Caldicellulosiruptor saccharolyticus Thesis Submitted to The School of Engineering of the UNIVERSITY OF DAYTON In Partial Fulfillment of the Requirements for The Degree of Master of Science in Chemical Engineering By Jin Cao Dayton, Ohio December, 2012 BIOCHEMICAL CHARACTERIZATION OF β-XYLAN ACTING GLYCOSIDE HYDROLASES FROM THE THERMOPHILIC BACTERIUM Caldicellulosiruptor saccharolyticus Name: Cao, Jin APPROVED BY: ______________________ ________________________ Donald A. Comfort, Ph.D. Amy Ciric, Ph.D. Advisory Committee Chairman Committee Member Research Advisor & Assistant Professor Lecturer Department of Department of Chemical & Materials Engineering Chemical & Materials Engineering ________________________ Karolyn Hansen, Ph.D. Committee Member Assistant Professor Department of Biology _______________________ ________________________ John G. Weber, Ph.D. Tony E. Saliba, Ph.D. Associate Dean Dean, School of Engineering School of Engineering & Wilke Distinguished Professor ii ABSTRACT BIOCHEMICAL CHARACTERIZATION OF Β-XYLAN ACTING GLYCOSIDE HYDROLASES FROM THE THERMOPHILIC BACTERIUM Caldicellulosiruptor saccharolyticus Name: Cao, Jin University of Dayton Research Advisor: Donald A. Comfort Fossil fuels have been the dominant source for energy around the world since the industrial revolution, however, with the increasing demand for energy and decreasing fossil fuel reserves alternative energy sources must be exploited. Bio-ethanol is a promising prospect for an alternative energy source to petroleum, especially when plant biomass is used as the replacement carbon source. This gives greater benefit for implementation of bioethanol as a viable alternative transport fuel than food stocks such as starch from corn. The implementation of this next generation of bioethanol requires more robust and environmentally friendly methods for degrading cellulose and hemicellulose, the two major components of plant biomass. -

Phylogenomic Analysis Supports the Ancestral Presence of LPS-Outer Membranes in the Firmicutes
Phylogenomic analysis supports the ancestral presence of LPS-outer membranes in the Firmicutes. Luisa Cs Antunes, Daniel Poppleton, Andreas Klingl, Alexis Criscuolo, Bruno Dupuy, Céline Brochier-Armanet, Christophe Beloin, Simonetta Gribaldo To cite this version: Luisa Cs Antunes, Daniel Poppleton, Andreas Klingl, Alexis Criscuolo, Bruno Dupuy, et al.. Phy- logenomic analysis supports the ancestral presence of LPS-outer membranes in the Firmicutes.. eLife, eLife Sciences Publication, 2016, 5, pp.e14589. 10.7554/eLife.14589.020. pasteur-01362343 HAL Id: pasteur-01362343 https://hal-pasteur.archives-ouvertes.fr/pasteur-01362343 Submitted on 8 Sep 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License RESEARCH ARTICLE Phylogenomic analysis supports the ancestral presence of LPS-outer membranes in the Firmicutes Luisa CS Antunes1†, Daniel Poppleton1†, Andreas Klingl2, Alexis Criscuolo3, Bruno Dupuy4, Ce´ line Brochier-Armanet5, Christophe Beloin6, Simonetta Gribaldo1* 1Unite´ de -
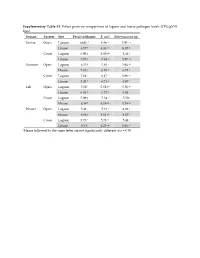
Supplementary Table S1. Fisher Pairwise Comparisons of Lagoon and House Pathogen Levels (CFU/Gvss Log10) Season System Site Fecal Coliforms E
Supplementary Table S1. Fisher pairwise comparisons of lagoon and house pathogen levels (CFU/gVSS log10) Season System Site Fecal coliforms E. coli Enterococcus sp. Spring Open Lagoon 6.00 g1 5.46 hi 5.91 cd House 6.57 e 6.33 cd 6.35 b Cover Lagoon 6.00 g 5.60 gh 5.44 f House 7.73 a 7.63 a 5.87 cd Summer Open Lagoon 6.35 f 5.83 f 5.94 cd House 7.83 a 6.10 e 6.73 a Cover Lagoon 7.04 c 6.47 c 5.86 cd House 7.42 b 6.71 b 6.07 c Fall Open Lagoon 5.58 i 5.58 gh 5.56 ef House 6.81 d 6.72 b 6.01 c Cover Lagoon 5.99 g 5.34 i 5.50 f House 6.38 f 6.19 de 5.74 de Winter Open Lagoon 5.41 j 5.13 j 4.88 g House 6.06 g 5.61 gh 6.67 a Cover Lagoon 5.73 h 5.73 fg 5.44 f House 6.34 f 6.25 de 5.86 cd 1Means followed by the same letter are not significantly different at p = 0.05 Supplementary Table S2. Relative abundances of OTUs identified using universal bacterial primer set, presented as relative abundances (%) Only bacterial families are counted in Figure 5 and discussion involving bacterial family identification. # Identified in # Identified from all 8 All 16 All #OTU Table SpOLRA SuOLRA FOLRA WOLRA SpCLRA SuCLRA FCLRA WCLRA SpOHRA SuOHRA FOHRA WOHRA SpCHRA SuCHRA FCHRA WCHRA all samples lagoon samples Samples Lagoons Notes Key Patulibacteraceae 0 9.93739E‐05* 0.000134 0.000116 0 0 0 0 0 0 0.000452 0 0 0 0 0 4 3 Sp = Spring O = Open Ruaniaceae 0 0 0 0 0 0 0 0 0 0 0 0 0 0.000221577 0 7.73E‐05 2 0 Su = Summer C = Covered Thermoactinomycetaceae 0 0 0.000267 0.000116 0 0.00037092 0.000529 0.000526 0 0.000342922 0.000271 0.000197 0 0 0 0 8 5 F = Fall L = Lagoon Beutenbergiaceae 0.000226334 0.000331246 0.000579 0.001663 0.000620176 0 0.000106 0.000807 0.002320743 0.000571537 0.000723 0.000591 0.000169731 0 0.000606 0 13 7 W = Winter H = House Thermoanaerobacterales Family III. -

Composition of Rabbit Caecal Microbiota and the Effects of Dietary Quercetin Supplementation and Sex Thereupon North M.K
W orld World Rabbit Sci. 2019, 27: 185-198 R abbit doi:10.4995/wrs.2019.11905 Science © WRSA, UPV, 2003 COMPOSITION OF RABBIT CAECAL MICROBIOTA AND THE EFFECTS OF DIETARY QUERCETIN SUPPLEMENTATION AND SEX THEREUPON NORTH M.K. *, DALLE ZOTTE A. †, HOFFMAN L.C. *‡ *Department of Animal Sciences, Stellenbosch University, Private Bag X1, Matieland, STELLENBOSCH 7602, South Africa. †Department of Animal Medicine, Production and Health, University of Padova, Agripolis, Viale dell’Università, 16, 35020 LEGNARO, Padova, Italy. ‡Centre for Nutrition and Food Sciences, Queensland Alliance for Agriculture and Food Innovation (QAAFI), The University of Queensland, Health and Food Sciences Precinct, 39 Kessels Road, COOPERS PLAINS 4108, Australia. Abstract: The purpose of this study was to add to the current understanding of rabbit caecal microbiota. This involved describing its microbial composition and linking this to live performance parameters, as well as determining the effects of dietary quercetin (Qrc) supplementation (2 g/kg feed) and sex on the microbial population. The weight gain and feed conversion ratio of twelve New Zealand White rabbits was measured from 5 to 12 wk old, blood was sampled at 11 wk old for the determination of serum hormone levels, and the rabbits were slaughtered and caecal samples collected at 13 wk old. Ion 16STM metagenome sequencing was used to determine the microbiota profile. The dominance of Firmicutes (72.01±1.14% of mapped reads), Lachnospiraceae (23.94±1.01%) and Ruminococcaceae (19.71±1.07%) concurred with previous reports, but variation both between studies and individual rabbits was apparent beyond this. Significant correlations between microbial families and live performance parameters were found, suggesting that further research into the mechanisms of these associations could be useful.