Gene Expression in Donor Corneal Endothelium
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MUC4/MUC16/Muc20high Signature As a Marker of Poor Prognostic for Pancreatic, Colon and Stomach Cancers
Jonckheere and Van Seuningen J Transl Med (2018) 16:259 https://doi.org/10.1186/s12967-018-1632-2 Journal of Translational Medicine RESEARCH Open Access Integrative analysis of the cancer genome atlas and cancer cell lines encyclopedia large‑scale genomic databases: MUC4/MUC16/ MUC20 signature is associated with poor survival in human carcinomas Nicolas Jonckheere* and Isabelle Van Seuningen* Abstract Background: MUC4 is a membrane-bound mucin that promotes carcinogenetic progression and is often proposed as a promising biomarker for various carcinomas. In this manuscript, we analyzed large scale genomic datasets in order to evaluate MUC4 expression, identify genes that are correlated with MUC4 and propose new signatures as a prognostic marker of epithelial cancers. Methods: Using cBioportal or SurvExpress tools, we studied MUC4 expression in large-scale genomic public datasets of human cancer (the cancer genome atlas, TCGA) and cancer cell line encyclopedia (CCLE). Results: We identifed 187 co-expressed genes for which the expression is correlated with MUC4 expression. Gene ontology analysis showed they are notably involved in cell adhesion, cell–cell junctions, glycosylation and cell signal- ing. In addition, we showed that MUC4 expression is correlated with MUC16 and MUC20, two other membrane-bound mucins. We showed that MUC4 expression is associated with a poorer overall survival in TCGA cancers with diferent localizations including pancreatic cancer, bladder cancer, colon cancer, lung adenocarcinoma, lung squamous adeno- carcinoma, skin cancer and stomach cancer. We showed that the combination of MUC4, MUC16 and MUC20 signature is associated with statistically signifcant reduced overall survival and increased hazard ratio in pancreatic, colon and stomach cancer. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Research2007herschkowitzetvolume Al
Open Access Research2007HerschkowitzetVolume al. 8, Issue 5, Article R76 Identification of conserved gene expression features between comment murine mammary carcinoma models and human breast tumors Jason I Herschkowitz¤*†, Karl Simin¤‡, Victor J Weigman§, Igor Mikaelian¶, Jerry Usary*¥, Zhiyuan Hu*¥, Karen E Rasmussen*¥, Laundette P Jones#, Shahin Assefnia#, Subhashini Chandrasekharan¥, Michael G Backlund†, Yuzhi Yin#, Andrey I Khramtsov**, Roy Bastein††, John Quackenbush††, Robert I Glazer#, Powel H Brown‡‡, Jeffrey E Green§§, Levy Kopelovich, reviews Priscilla A Furth#, Juan P Palazzo, Olufunmilayo I Olopade, Philip S Bernard††, Gary A Churchill¶, Terry Van Dyke*¥ and Charles M Perou*¥ Addresses: *Lineberger Comprehensive Cancer Center. †Curriculum in Genetics and Molecular Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA. ‡Department of Cancer Biology, University of Massachusetts Medical School, Worcester, MA 01605, USA. reports §Department of Biology and Program in Bioinformatics and Computational Biology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA. ¶The Jackson Laboratory, Bar Harbor, ME 04609, USA. ¥Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA. #Department of Oncology, Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC 20057, USA. **Department of Pathology, University of Chicago, Chicago, IL 60637, USA. ††Department of Pathology, University of Utah School of Medicine, Salt Lake City, UT 84132, USA. ‡‡Baylor College of Medicine, Houston, TX 77030, USA. §§Transgenic Oncogenesis Group, Laboratory of Cancer Biology and Genetics. Chemoprevention Agent Development Research Group, National Cancer Institute, Bethesda, MD 20892, USA. Department of Pathology, Thomas Jefferson University, Philadelphia, PA 19107, USA. Section of Hematology/Oncology, Department of Medicine, Committees on Genetics and Cancer Biology, University of Chicago, Chicago, IL 60637, USA. -

NIH Public Access Author Manuscript Dev Biol
NIH Public Access Author Manuscript Dev Biol. Author manuscript; available in PMC 2013 January 1. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Dev Biol. 2012 January 1; 361(1): 68±78. doi:10.1016/j.ydbio.2011.10.004. Regulation of intrahepatic biliary duct morphogenesis by Claudin 15-like b Isla D. Cheung1, Michel Bagnat1,2, Taylur P. Ma3, Anirban Datta4, Kimberley Evason1,5, John C. Moore6, Nathan Lawson6, Keith E. Mostov4, Cecilia B. Moens3, and Didier Y.R. Stainier1,* 1Department of Biochemistry and Biophysics; Programs in Developmental and Stem Cell Biology, Genetics and Human Genetics; Liver Center, Diabetes Center, and Institute for Regeneration Medicine; University of California, San Francisco, CA 94158, USA 3HHMI and Division of Basic Science, Fred Hutchinson Cancer Research Center, Seattle WA 98109, USA 4Department of Anatomy, University of California, San Francisco, CA 94143, USA 5Department of Pathology, University of California, San Francisco, CA 94110, USA 6Program in Gene Function and Expression, University of Massachusetts Medical School, Worcester, MA 01605, USA Abstract The intrahepatic biliary ducts transport bile produced by the hepatocytes out of the liver. Defects in biliary cell differentiation and biliary duct remodeling cause a variety of congenital diseases including Alagille Syndrome and polycystic liver disease. While the molecular pathways regulating biliary cell differentiation have received increasing attention (Lemaigre, 2010), less is known about the cellular behavior underlying biliary duct remodeling. Here, we have identified a novel gene, claudin 15-like b (cldn15lb), which exhibits a unique and dynamic expression pattern in the hepatocytes and biliary epithelial cells in zebrafish. -

Novel Compound Heterozygote Mutations of TJP2 in a Chinese
Ge et al. BMC Medical Genetics (2019) 20:18 https://doi.org/10.1186/s12881-019-0753-7 CASEREPORT Open Access Novel compound heterozygote mutations of TJP2 in a Chinese child with progressive cholestatic liver disease Ting Ge, Xinyue Zhang, Yongmei Xiao, Yizhong Wang* and Ting Zhang* Abstract Background: Progressive familial intrahepatic cholestasis (PFIC) is a group of genetic autosomal recessive disorders that predominantly affects young children and results in early-onset progressive liver damage. Several types of PFIC were defined based on different genetic aetiologies in last decades. Case presentation: Here, we report a Chinese young child diagnosed as PFIC with variants in tight junction protein 2 (TJP2). The patient was affected by a long history of jaundice, pruritus, and failure to thrive. Highly elevated level of serum total bile acid (TBA) and normal levels of gamma-glutamyltransferase (GGT) were observed at hospitalization. The patient’s clinical symptoms could be alleviated by administration of ursodeoxycholic acid. Genetic testing by next generation sequencing (NGS) found novel compound heterozygote mutations c.2448 + 1G > C/c.2639delC (p.T880Sfs*12) in TJP2, which were inherited from her mother and father, respectively. Both mutations were predicted to abolish TJP2 protein translation, and neither has previously been identified. Conclusion: We report a Chinese female PFIC child with novel compound heterozygous mutations of TJP2.Genetic testing by NGS is valuable in the clinical diagnosis of hereditary liver disease. Keywords: Autosomal recessive disorder, Child, Compound heterozygote mutations, Progressive cholestatic liver disease, TJP2 Background in adenosine triphosphate-binding cassette subfamily B Progressive familial intrahepatic cholestasis (PFIC), first de- member 4 (ABCB4), which encodes the multidrug scribed in 1969, is a group of genetic autosomal recessive resistance P-glycoprotein 3 protein (MDR3) leads to disorders that are characterized by fluctuating jaundice, the development of PFIC 3 [5]. -

A Novel Homozygous USP53 Splicing Variant Disrupting the Gene Function That Causes Cholestasis Phenotype and Review of the Literature
A Novel Homozygous USP53 Splicing Variant Disrupting the Gene Function that Causes Cholestasis Phenotype and Review of the Literature ALPER GEZDIRICI ( [email protected] ) Basaksehir Cam and Sakura City Hospital https://orcid.org/0000-0002-2432-9279 ÖZLEM KALAYCIK ŞENGÜL İstanbul Kanuni Sultan Süleyman Eğitim ve Araştırma Hastanesi: Istanbul Kanuni Sultan Suleyman Egitim ve Arastirma Hastanesi MUSTAFA DOĞAN Basaksehir Cam and Sakura City Hospital BANU YILMAZ ÖZGÜVEN Basaksehir Cam and Sakura City Hospital EKREM AKBULUT Malatya Turgut Özal Üniversitesi: Malatya Turgut Ozal Universitesi Research Article Keywords: USP53, cholestasis, homology modeling, whole exome sequencing Posted Date: August 12th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-762230/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/19 Abstract Background: Hereditary cholestasis is a heterogeneous group of liver diseases that mostly show autosomal recessive inheritance. The phenotype of cholestasis is highly variable. Molecular genetic testing offers a useful approach to differentiate different types of cholestasis because some symptoms and ndings overlap. Biallelic variants in USP53 have recently been reported in cholestasis phenotype. In this study we aim to characterize clinical ndings and biological insights on a novel USP53 splice variant causing cholestasis phenotype and review of the literature. Methods and Results: We reported a novel splice variant (NM_019050.2:c.238-1G>C) in the USP53 gene via whole exome sequencing in a patient with cholestasis phenotype. This variant was conrmed by sanger sequencing and as a result of family segregation analysis; it was found to be a heterozygous state in the parents and the other healthy elder brother of our patient. -

Protective Effects of Propionate Upon the Blood–Brain Barrier
bioRxiv preprint doi: https://doi.org/10.1101/170548; this version posted February 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Microbiome–host systems interactions: Protective effects of propionate upon 2 the blood–brain barrier 3 4 Lesley Hoyles1*, Tom Snelling1, Umm-Kulthum Umlai1, Jeremy K. Nicholson1, Simon 5 R. Carding2, 3, Robert C. Glen1, 4 & Simon McArthur5* 6 7 1Division of Integrative Systems Medicine and Digestive Disease, Department of 8 Surgery and Cancer, Imperial College London, UK 9 2Norwich Medical School, University of East Anglia, UK 10 3The Gut Health and Food Safety Research Programme, The Quadram Institute, 11 Norwich Research Park, Norwich, UK 12 4Centre for Molecular Informatics, Department of Chemistry, University of Cambridge, 13 Cambridge, UK 14 5Institute of Dentistry, Barts & the London School of Medicine & Dentistry, Blizard 15 Institute, Queen Mary University of London, London, UK 16 17 *Corresponding authors: Lesley Hoyles, [email protected]; Simon 18 McArthur, [email protected] 19 Running title: Propionate affects the blood–brain barrier 20 Abbreviations: ADHD, attention-deficit hyperactivity disorder; ASD, autism spectrum 21 disorder; BBB, blood–brain barrier; CNS, central nervous system; FFAR, free fatty acid 22 receptor; KEGG, Kyoto Encyclopaedia of Genes and Genomes; GO, Gene Ontology; 1 bioRxiv preprint doi: https://doi.org/10.1101/170548; this version posted February 7, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Novel Protein Pathways in Development and Progression of Pulmonary Sarcoidosis Maneesh Bhargava1*, K
www.nature.com/scientificreports OPEN Novel protein pathways in development and progression of pulmonary sarcoidosis Maneesh Bhargava1*, K. J. Viken1, B. Barkes2, T. J. Grifn3, M. Gillespie2, P. D. Jagtap3, R. Sajulga3, E. J. Peterson4, H. E. Dincer1, L. Li2, C. I. Restrepo2, B. P. O’Connor5, T. E. Fingerlin5, D. M. Perlman1 & L. A. Maier2 Pulmonary involvement occurs in up to 95% of sarcoidosis cases. In this pilot study, we examine lung compartment-specifc protein expression to identify pathways linked to development and progression of pulmonary sarcoidosis. We characterized bronchoalveolar lavage (BAL) cells and fuid (BALF) proteins in recently diagnosed sarcoidosis cases. We identifed 4,306 proteins in BAL cells, of which 272 proteins were diferentially expressed in sarcoidosis compared to controls. These proteins map to novel pathways such as integrin-linked kinase and IL-8 signaling and previously implicated pathways in sarcoidosis, including phagosome maturation, clathrin-mediated endocytic signaling and redox balance. In the BALF, the diferentially expressed proteins map to several pathways identifed in the BAL cells. The diferentially expressed BALF proteins also map to aryl hydrocarbon signaling, communication between innate and adaptive immune response, integrin, PTEN and phospholipase C signaling, serotonin and tryptophan metabolism, autophagy, and B cell receptor signaling. Additional pathways that were diferent between progressive and non-progressive sarcoidosis in the BALF included CD28 signaling and PFKFB4 signaling. Our studies demonstrate the power of contemporary proteomics to reveal novel mechanisms operational in sarcoidosis. Application of our workfows in well-phenotyped large cohorts maybe benefcial to identify biomarkers for diagnosis and prognosis and therapeutically tenable molecular mechanisms. -

View: Regulation of Epithelial Na Channel Traffick- Ing
BASIC RESEARCH www.jasn.org Phosphoproteomic Profiling Reveals Vasopressin- Regulated Phosphorylation Sites in Collecting Duct Amar D. Bansal,* Jason D. Hoffert,* Trairak Pisitkun,* Shelly Hwang,* Chung-Lin Chou,* Emily S. Boja,† Guanghui Wang,† and Mark A. Knepper* *Epithelial Systems Biology Laboratory, and †Proteomics Core Facility, Division of Intramural Research, National Heart, Lung and Blood Institute, National Institutes of Health, Bethesda, Maryland ABSTRACT Protein phosphorylation is an important component of vasopressin signaling in the renal collecting duct, but the database of known phosphoproteins is incomplete. We used tandem mass spectrometry to identify vasopressin-regulated phosphorylation events in isolated rat inner medullary collecting duct (IMCD) suspensions. Using multiple search algorithms to identify the phosphopeptides from spectral data, we expanded the size of the existing collecting duct phosphoproteome database from 367 to 1187 entries. Label-free quantification in vasopressin- and vehicle-treated samples detected a significant change in the phosphorylation of 29 of 530 quantified phosphopeptides. The targets include important structural, regulatory, and transporter proteins. The vasopressin-regulated sites included two known sites (Ser-486 and Ser-499) present in the urea channel UT-A1 and one previously unknown site (Ser-84) on vasopressin-sensitive urea channels UT-A1 and UT-A3. In vitro assays using synthetic peptides showed that purified protein kinase A (PKA) could phosphorylate all three sites, and immunoblotting confirmed the PKA dependence of Ser-84 and Ser-486 phosphorylation. These results expand the known list of collecting duct phosphoproteins and highlight the utility of targeted phosphoproteomic approaches. J Am Soc Nephrol 21: 303–315, 2010. doi: 10.1681/ASN.2009070728 Vasopressin plays a central role in collecting duct In a previous study,17 we used tandem mass physiology. -

Effect of Graphene and Graphene Oxide on Airway Barrier and Differential Phosphorylation of Proteins in Tight and Adherens Junction Pathways
nanomaterials Article Effect of Graphene and Graphene Oxide on Airway Barrier and Differential Phosphorylation of Proteins in Tight and Adherens Junction Pathways Sofie Van Den Broucke 1, Jeroen A. J. Vanoirbeek 2 , Rita Derua 3, Peter H. M. Hoet 1,2,* and Manosij Ghosh 1,2,* 1 Centre for Environment and Health, Department of Public Health and Primary Care, KU Leuven, 3000 Leuven, Belgium; vandenbroucke.sofi[email protected] 2 Laboratory of Respiratory Diseases and Thoracic Surgery (BREATHE), KU Leuven, 3000 Leuven, Belgium; [email protected] 3 Laboratory of Protein Phosphorylation and Proteomics, KU Leuven, 3000 Leuven, Belgium; [email protected] * Correspondence: [email protected] (P.H.M.H.); [email protected] (M.G.) Abstract: Via inhalation we are continuously exposed to environmental and occupational irritants which can induce adverse health effects, such as irritant-induced asthma (IIA). The airway epithelium forms the first barrier encountered by these agents. We investigated the effect of environmental and occupational irritants on the airway epithelial barrier in vitro. The airway epithelial barrier was mimicked using a coculture model, consisting of bronchial epithelial cells (16HBE) and monocytes (THP-1) seeded on the apical side of a permeable support, and human lung microvascular endothelial cells (HLMVEC) grown on the basal side. Upon exposure to graphene (G) and graphene oxide Citation: Van Den Broucke, S.; (GO) in a suspension with fetal calf serum (FCS), ammonium persulfate (AP), sodium persulfate Vanoirbeek, J.A.J.; Derua, R.; Hoet, (SP) and hypochlorite (ClO−), the transepithelial electrical resistance (TEER) and flux of fluorescent P.H.M.; Ghosh, M. -

S12920-021-01005-X.Pdf
Tkatchenko and Tkatchenko BMC Med Genomics (2021) 14:153 https://doi.org/10.1186/s12920-021-01005-x RESEARCH Open Access Genome-wide analysis of retinal transcriptome reveals common genetic network underlying perception of contrast and optical defocus detection Tatiana V. Tkatchenko1 and Andrei V. Tkatchenko1,2,3* Abstract Background: Refractive eye development is regulated by optical defocus in a process of emmetropization. Excessive exposure to negative optical defocus often leads to the development of myopia. However, it is still largely unknown how optical defocus is detected by the retina. Methods: Here, we used genome-wide RNA-sequencing to conduct analysis of the retinal gene expression network underlying contrast perception and refractive eye development. Results: We report that the genetic network subserving contrast perception plays an important role in optical defo- cus detection and emmetropization. Our results demonstrate an interaction between contrast perception, the retinal circadian clock pathway and the signaling pathway underlying optical defocus detection. We also observe that the relative majority of genes causing human myopia are involved in the processing of optical defocus. Conclusions: Together, our results support the hypothesis that optical defocus is perceived by the retina using con- trast as a proxy and provide new insights into molecular signaling underlying refractive eye development. Keywords: Refractive eye development, Myopia, Contrast perception, Optical defocus, Circadian rhythms, Signaling pathways, Gene expression profling, Genetic network, RNA-seq Background compensate for imposed defocus very accurately by Refractive eye development is controlled by both envi- modulating the growth of the posterior segment of the ronmental and genetic factors, which determine the eye via a developmental mechanism called bidirectional optical geometry of the eye and its refractive state by a emmetropization by the sign of optical defocus (BESOD), process called emmetropization [1–8]. -
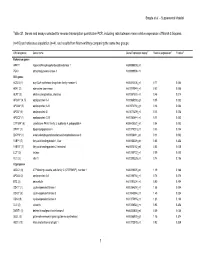
Table S1. Genes and Assays Selected for Reverse Transcription Quantitative PCR, Including Ratio Between Mean Relative Expression of Marsh 3 Biopsies
Bragde et al. – Supplemental Material Table S1. Genes and assays selected for reverse transcription quantitative PCR, including ratio between mean relative expression of Marsh 3 biopsies (n=10) and reference population (n=4), and results from Mann-whitney comparing the same two groups. Official symbol Gene name Gene Expression assay** Relative expression‡ P-value§ Reference genes HPRT1 hypoxanthine phosphoribosyltransferase 1 Hs99999909_m1 PGK1 phosphoglycerate kinase 1 Hs99999906_m1 Villi genes ACSL5 (1) acyl-CoA synthetase long-chain family member 5 Hs00212106_m1 0.77 0.054 ADA* (2) adenosine deaminase Hs01110949_m1 0.52 0.008 ALPI* (3) alkaline phosphatase, intestinal Hs00357578_m1 0.46 0.014 APOA1* (4, 5) apolipoprotein A-I Hs00985000_g1 0.05 0.002 APOA4* (5) apolipoprotein A-IV Hs01573751_g1 0.16 0.002 APOB* (5) apolipoprotein B Hs01071209_m1 0.13 0.002 APOC3* (1) apolipoprotein C-III Hs00163644_m1 0.11 0.002 CYP3A4* (6) cytochrome P450, family 3, subfamily A, polypeptide 4 Hs00430021_m1 0.04 0.002 DPP4* (1) dipeptidyl-peptidase 4 Hs00175210_m1 0.33 0.004 ENPP3* (1) ectonucleotide pyrophosphatase/phosphodiesterase 3 Hs01038411_m1 0.11 0.002 FABP1 (7) fatty acid binding protein 1, liver Hs00155026_m1 0.85 0.454 FABP2* (7) fatty acid binding protein 2, intestinal Hs01573162_m1 0.53 0.008 LCT* (2) lactase Hs00158722_m1 0.09 0.002 VIL1 (2) villin 1 Hs00200229_m1 0.74 0.106 Crypt genes ABCC1 (8) ATP-binding cassette, sub-family C (CFTR/MRP), member 1 Hs00219905_m1 1.19 0.188 APOA2 (2) apolipoprotein A-II Hs00155788_m1 0.78 0.076 BTC (2) betacellulin Hs01101204_m1 0.90 0.454 CDK1* (1) cyclin-dependent kinase 1 Hs00364293_m1 1.58 0.004 CDK2* (9) cyclin-dependent kinase 2 Hs01548894_m1 1.45 0.024 CDK4 (9) cyclin-dependent kinase 4 Hs00175935_m1 1.23 0.188 CLU (2) clusterin Hs00156548_m1 0.93 0.454 DMBT1 (1) deleted in malignant brain tumors 1 Hs00244838_m1 0.69 0.054 GLUL (2) glutamate-ammonia ligase (glutamine synthetase) Hs00365928_g1 1.15 0.374 HES1 (10) Hairy and enhancer of split 1 Hs00172878_m1 0.92 0.839 1 Bragde et al.