Shallow Population Genetic Structures of Thread-Sail Filefish (Stephanolepis Cirrhifer) Populations from Korean Coastal Waters
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Teeth and Dentition of the Filefish (Stephanolepis Cirrhifer) Revisited Tomographically
1 J-STAGE Advance Publication: August 12, 2020 Journal of Oral Science Original article The teeth and dentition of the filefish (Stephanolepis cirrhifer) revisited tomographically Hirofumi Kanazawa1,2), Maki Yuguchi1,2,3), Yosuke Yamazaki1,2,3), and Keitaro Isokawa1,2,3) 1) Division of Oral Structural and Functional Biology, Nihon University Graduate School of Dentistry, Tokyo, Japan 2) Department of Anatomy, Nihon University School of Dentistry, Tokyo, Japan 3) Division of Functional Morphology, Dental Research Center, Nihon University School of Dentistry, Tokyo, Japan (Received October 31, 2019; Accepted November 26, 2019) Abstract: The upper and lower tooth-bearing jaws of the filefish (Stepha- Teeth of Non-Mammalian Vertebrates, Elsevier, 2017). nolepis cirrhifer) were scanned using a micro-CT system in order to With regard to filefish dentition in Japan, an early morphologic and address the existing gaps between the traditional pictures of the morphol- histologic study of Monocanthus cirrhifer and Cantherines modestus ogy and histology. 2D tomograms, reconstructed 3D models and virtual (synonyms Stephanolepis cirrhifer and Thamnaconus modestus, respec- dissection were employed to examine and evaluate the in situ geometry of tively) was carried out by Sohiti Isokawa (Isokawa, Zool Mag 64, 194-197, tooth implantation and the mode of tooth attachment both separately and 1955). Phylogenetic interrelationships in the balistoids were examined collectively. No distinct sockets comparable to those in mammals were extensively by Matsuura [4], based on many anatomical characteristics evident, but shallow depressions were observed in the premaxillary and including the tooth-bearing jaws, which were the premaxillary and the the dentary. The opening of the tooth pulp cavity was not simply oriented dentary. -

Title Laboratory Video Recordings and Underwater Visual Observations
Laboratory video recordings and underwater visual Title observations combined to reveal activity rhythm of red-spotted grouper and banded wrasse, and their natural assemblages Author(s) Masuda, Reiji; Matsuda, Katsuhiro; Tanaka, Masaru Citation Environmental Biology of Fishes (2012), 95(3): 335-346 Issue Date 2012-11 URL http://hdl.handle.net/2433/160388 Right The final publication is available at www.springerlink.com Type Journal Article Textversion author Kyoto University 1 Laboratory video recordings and underwater visual observations combined to reveal activity rhythm of 2 red-spotted grouper and banded wrasse, and their natural assemblages 3 4 Reiji Masuda, Katsuhiro Matsuda, Masaru Tanaka 5 6 7 8 R. Masuda 9 Maizuru Fisheries Research Station, Kyoto University, Nagahama, Maizuru, Kyoto 6250086, Japan 10 e-mail: [email protected] 11 Telephone: +81-773-62-9063, Fax: +81-773-62-5513 12 13 K. Matsuda 14 Kyushu Branch, Ajinomoto Co., Inc., 3-3-31 Ekiminami, Hakata, Fukuoka 8128683, Japan 15 16 M. Tanaka 17 International Institute for Advanced Studies, 9-3 Kizugawadai, Kizugawa-shi, Kyoto 6190225, Japan 1 18 Abstract The diel activity rhythm of red-spotted grouper Epinephelus akaara was studied both in 19 captivity and in the wild. Behavior of solitary grouper (58 to 397 mm in total length) in a tank was video 20 recorded using infrared illuminators under 11L/10D and two 1.5-h twilight transition periods, and was 21 compared to that of banded wrasse Halichoeres poecilopterus, a typical diurnal fish. Underwater 22 observations using SCUBA were also conducted in their natural habitat to reveal the behavioral activity 23 together with a visual census of adjacent fish and crustacean assemblages. -

Vertical Distribution and Feeding Ecology of the Black Scraper, Thamnaconus Modestus, in the Southern Sea of Korea
www.trjfas.org ISSN 1303-2712 Turkish Journal of Fisheries and Aquatic Sciences 13: 249-259 (2013) DOI: 10.4194/1303-2712-v13_2_07 Vertical Distribution and Feeding Ecology of the Black Scraper, Thamnaconus modestus, in the Southern Sea of Korea Hye Rim Kim1, Jung Hwa Choi1,*, Won Gyu Park2 1 National Fisheries Research and Development Institute, Busan 619-705, Korea. 2 Pukyong National University, Department of Marine Biology, , Busan 608-737, Korea. * Corresponding Author: Tel.: 82-51-270-2291; Fax: 82-51-270-2291; Received 02 October 2012 E-mail: [email protected] Accepted 01 April 2013 Abstract We investigated the vertical distribution and feeding ecology of the black scraper, Tamnaconus modestus, in the southern sea of Korea in the spring of 2009, 2010 and 2011 using otter trawl for 60 min per site. Fish abundance was significantly related to the basis of depth and temperature. Thamnaconus modestus occurred at depths ranging from 80 to 120 meter waters having temperature higher than 12°C. We found that 87% of the total catch were obtained at the depths between 80 and 120 meters (58% and 29% from 80–100 and 100–120 meters respectively). The total length of captured individuals ranged from 10.6 to 38.7 cm. Individuals captured at deeper were significantly larger than those from shallower sites. Diverse prey organisms, including algae, amphipods, gastropods, ophiuroids, and cephalopods, were found in T. modestus stomach. The main prey items in IRI value belonged to four groups: hyperiid amphipods, gastropods, ophiuroids, and algae. The fish showed ontogenetic diet shifts. In general, individuals in smaller size (< 20 cm) shifted their diets from vegetative food sources to animal ones when they reach the size bigger than 20 cm. -

Parasitic Copepods of Marine Fish Cultured in Japan: a Review Kazuya Nagasawa*
Journal of Natural History, 2015 Vol. 49, Nos. 45–48, 2891–2903, http://dx.doi.org/10.1080/00222933.2015.1022615 Parasitic copepods of marine fish cultured in Japan: a review Kazuya Nagasawa* Graduate School of Biosphere Science, Hiroshima University, Hiroshima, Japan (Received 22 September 2014; accepted 4 February 2015; first published online 29 June 2015) This paper reviews aspects of the biology of copepods infecting marine fish commer- cially cultured at fish farms or held as broodstock at governmental hatcheries in Japan. In total, 20 species of parasitic copepods have been reported from these fish: they are mostly caligids (12 spp.), followed by lernaeopodids (4 spp.), pennellid (1 sp.), chondracanthid (1 sp.), taeniacanthid (1 sp.), and unidentified species (1 sp.). The identified copepods are: Caligus fugu, C. lagocephalus, C. lalandei, C. latigenitalis, C. longipedis, C. macarovi, C. orientalis, C. sclerotinosus, C. spinosus, Lepeophtheirus longiventralis, L. paralichthydis, L. salmonis (Caligidae); Alella macrotrachelus, Clavella parva, Parabrachiella hugu, P. seriolae (Lernaeopodidae); Peniculus minuti- caudae (Pennellidae); Acanthochondria priacanthi (Chondracanthidae); and Biacanthus pleuronichthydis (Taeniacanthidae). The fish recorded as hosts include carangids (4 spp.), sparids (2 spp.), monacanthids (2 spp.), salmonids (2 spp.), scom- brid (1 sp.), tetraodontid (1 sp.), pleuronectid (1 sp.), paralichthyid (1 sp.), and trichodontid (1 sp.). Only five species (C. orientalis, L. longiventralis, L. salmonis, C. parva and A. priacanthi) parasitize farmed fish in subarctic waters, while all other species (15 spp.) infect farmed fish in temperate waters. No information is yet avail- able on copepods from fish farmed in subtropical waters. Three species of Caligus (C. fugu, C. sclerotinosus and C. -

On the Species of the Genus Balistes Described by Johann Julius Walbaum (1792)
ON THE SPECIES OF THE GENUS BALISTES DESCRIBED BY JOHANN JULIUS WALBAUM (1792) by Paolo PARENTI (1) ABSTRACT. - The status of six nominal species placed in the genus Balistes by Johann J. Walbaum (1792) is reported. Balistes longirostris and B. meulenii are shown to be senior synonyms of Oxymonacanthus longirostris (Bloch & Schneider, 1801) and Cantherines fronticinctus (Günther, 1867), respectively. Conditions exist, however, that allow “pre- vailing usage” of the latter names, as provided by Article 23.9.1 of the International Code of Zoological Nomenclature. Balistes auwawa, B. barbatus and B. talpa are shown to be junior synonyms of Aluterus monoceros (Linnaeus, 1758), whereas B. capriscus is a junior homonym of B. capriscus Gmelin, 1789 and therefore objectively invalid. RÉSUMÉ. - Sur les espèces du genre Balistes décrites par Johann Julius Walbaum (1792). Le statut de six taxons nominaux placés parmi le genre Balistes par Johann J. Walbaum (1792) a été déterminé. Balistes longirostris et B. meulenii sont des synonymes seniors d’Oxymonacanthus longirostris (Bloch & Schneider, 1801) et de Cantherines fronticinctus (Günther, 1867), respectivement. Toutefois, les conditions existent pour maintenir l’usage prédominant de ces derniers, d’après l’Article 23.9.1 du Code International de Nomenclature Zoologique. B. auwawa, B. barbatus et B. talpa sont tous des synonymes juniors d’Aluterus monoceros (Linnaeus, 1758), tandis que B. capriscus est un homonyme junior de B. capriscus Gmelin, 1789 et, pour cette raison, non valide. Key words. - Monacanthidae - Balistes - Taxonomy - Senior synonyms - Nomen protectum - Nomen oblitum - Walbaum. Johann Julius Walbaum (1724 - 1799) (Fig. 1), doctor of amination of the large group of unplaced nominal species medicine in Lubeck and owner of a cabinet of natural revealed that most of these represent senior or junior syn- objects (destroyed during World War II), became well onyms of well known fish species (Parenti, 2002; Parenti known through several zoological publications (see Müller, and Pietsch, 2002). -

Fish Assemblages Associated with Three Types of Artificial Reefs: Density of Assemblages and Possible Impacts on Adjacent Fish Abundance
Fish assemblages associated with three types of artificial reefs: density of assemblages and possible impacts on adjacent fish abundance Item Type article Authors Masuda, Reiji; Shiba, Masami; Yamashita, Yoh; Ueno, Masahiro; Kai, Yoshiaki; Nakanishi, Asami; Torikoshi, Masaru; Tanaka, Masaru Download date 26/09/2021 23:03:05 Link to Item http://hdl.handle.net/1834/25404 162 Abstract—We evaluated the effective- ness of wooden artificial reefs (ARs) Fish assemblages associated as fish habitat. Three types of ARs, with three types of artificial reefs: made of cedar logs, broadleaf tree logs, and PVC pipes, respectively, density of assemblages were deployed in triplicate at 8-m and possible impacts depth off Maizuru, Kyoto Prefec- ture, Sea of Japan, in May 2004. Fish on adjacent fish abundance assemblages associated with each of the nine ARs were observed by using 1 1 SCUBA twice a month for four years. Reiji Masuda (contact author) Yoshiaki Kai Fish assemblages in the adjacent Masami Shiba2 Asami Nakanishi1 habitat were also monitored for two 1 1 years before and four years after reef Yoh Yamashita Masaru Torikoshi deployment. In the surveyed areas Masahiro Ueno1 Masaru Tanaka3 (ca. 10 m2) associated with each of the Email address for contact author: [email protected] cedar, broadleaf, and PVC ARs, the average number of fish species was 1 Maizuru Fisheries Research Station 4.14, 3.49, and 3.00, and the average Kyoto University number of individuals was 40.7, 27.9, Nagahama, Maizuru and 20.3, respectively. The estimated Kyoto 625-0086, Japan biomass was also more greater when 2 Ashiu Forest Research Station associated with the cedar ARs than Kyoto University with other ARs. -

Redalyc.Feeding Ecology of the Planehead Filefish Stephanolepis
Revista de Biología Marina y Oceanografía ISSN: 0717-3326 [email protected] Universidad de Valparaíso Chile Mancera-Rodríguez, Néstor Javier; Castro-Hernández, José Juan Feeding ecology of the planehead filefish Stephanolepis hispidus (Pisces: Monacanthidae), in the Canary Islands area Revista de Biología Marina y Oceanografía, vol. 50, núm. 2, agosto, 2015, pp. 221-234 Universidad de Valparaíso Viña del Mar, Chile Available in: http://www.redalyc.org/articulo.oa?id=47941332002 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative Revista de Biología Marina y Oceanografía Vol. 50, Nº2: 221-234, agosto 2015 DOI 10.4067/S0718-19572015000300002 ARTICLE Feeding ecology of the planehead filefish Stephanolepis hispidus (Pisces: Monacanthidae), in the Canary Islands area Ecología trófica de Stephanolepis hispidus (Pisces: Monacanthidae), en el área de las Islas Canarias Néstor Javier Mancera-Rodríguez1 and José Juan Castro-Hernández2 1Universidad Nacional de Colombia, Department of Forestry Sciences, Calle 59A No. 63-20, Bloque 20, oficina 211, Medellín, Colombia. [email protected] 2Universidad de Las Palmas de Gran Canaria, Department of Biology, Edf. Ciencias Básicas, Campus de Tafira, 35017, Las Palmas de Gran Canaria, Islas Canarias, España. [email protected] Resumen.- Se examinó la ecología alimentaria de Stephanolepis hispidus, en aguas de las Islas Canarias. El estudio se basó en el contenido estomacal de 823 ejemplares de 8,9 cm a 25,9 cm de longitud total (LT), capturados mensualmente en trampas para peces entre febrero de 1998 y junio de 1999. -
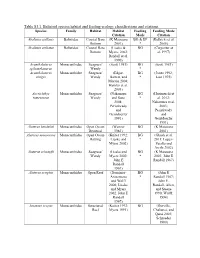
Table S3.1. Balistoid Species Habitat and Feeding Ecology Classifications and Citations
Table S3.1. Balistoid species habitat and feeding ecology classifications and citations. Species Family Habitat Habitat Feeding Feeding Mode Citation Mode Citation Abalistes stellaris Balistidae Coastal Bare (K Matsuura BG & EP (Kulbicki et al. Bottom 2001) * 2005) Abalistes stellatus Balistidae Coastal Bare (Lieske & BG (Carpenter et Bottom Myers, 2002; al. 1997) Randall et al. 1990) Acanthaluteres Monacanthidae Seagrass/ (Scott 1981) BG (Scott 1981) spilomelanurus Weedy * Acanthaluteres Monacanthidae Seagrass/ (Edgar, BG (Jones 1992; vittiger Weedy Barrett, and * Last 1975) Morton 2004; Hyndes et al. 2003) Acreichthys Monacanthidae Seagrass/ (Nakamura BG (Horinouchi et tomentosus Weedy and Sano * al. 2012; 2004; Nakamura et al. Peristiwady 2003; and Peristiwady Geistdoerfer and 1991) Geistdoerfer 1991) Aluterus heudeloti Monacanthidae Open Ocean (Wenner BG (K Matsuura Demersal 1983) 2001) Aluterus monoceros Monacanthidae Open Ocean (Kuiter 1992; BG (Ghosh et al. Rafting Lieske and * 2011; Lopez- Myers 2002) Peralta and Arcila 2002) Aluterus schoepfii Monacanthidae Seagrass/ (Lieske and BG (K Matsuura Weedy Myers 2002; * 2001; John E John E Randall 1967) Randall 1967) Aluterus scriptus Monacanthidae Open Reef (Dominici- BG (John E Arosemena * Randall 1967; and Wolff John E. 2006; Lieske Randall, Allen, and Myers and Steene 2002; John E 1990; Wulff, Randall 1994) 1967) Amanses scopas Monacanthidae Structured (Kuiter 1992; BG (Durville, Reef Myers 1991) Chabanet, and Quod 2003; Schroeder 1980) Balistapus Balistidae Structured (Hiatt and BG & EP (Hiatt and undulatus Reef Strasburg * Strasburg 1960; 1960; Lieske John E. Randall and Myers 1955; John E. 2002; Randall, Allen, McClanahan and Steene and Shafir 1990) 1990; John E. Randall, Allen, and Steene 1990) Balistes capriscus Balistidae Open Ocean (Longley BG & EP (Goldman, Pelagic 1941) * Glasgow, and Falk 2016; Lieske and Myers 2002; Vose and Nelson 1994) Balistes polylepis Balistidae Open Reef (Burgess et BG & EP (Abitia al. -

Diel Changes in the Diet of Rudarius Ercodes: a Diurnal Omnivore and Nocturnal Carnivore
Korean J. Ichthyol. 18(3), 178~183, 2006 Diel Changes in the Diet of Rudarius ercodes: A Diurnal Omnivore and Nocturnal Carnivore Seok Nam Kwak*, Sung-Hoi Huh1 and Chang Geun Choi2 Korea Inter-university Institutes of Ocean Science, and 1Department of Oceanography, Pukyong National University, Busan 608-737, Korea 2Research Institute of Marine Science and Technology, Korea Maritime University, Busan 606-791, Korea Diel changes in the feeding habits of Rudarius ercodes were investigated in an eelgrass bed of Jindong Bay, Korea. The main food components for R. ercodes (1.6~4.3 cm SL) were gammarid amphipods, eelgrass, polychaetes and urochordates. Most dietary items were inhabitants of an eelgrass bed. Diel variations in diet and feeding activity occurred. The diet of R. ercodes underwent changes from eelgrass and gammarid amphipods (omnivore) at day to mainly gammarid amphipods, polychaetes, and urochordates (carnivore) at night. Rudarius ercodes probably took detached eelgrass leaves and grazed live eelgrass during day, whereas feeding on gammarid amphipods, polychaetes, urochordates, and bivalves were facilitated by nocturnal movement and activity of these prey during night. The feeding activity of R. ercodes was also correlated with periods of high tides. Key words : Rudarius ercodes, diel, diet, gammarid amphipods, eelgrass, poly- chaetes, urochordates, tide cirrhifer, suggested that it is a omnivore, feeding Introduction primarily amphipods (gammarids and caprellids) and eelgrass (Kwak et al., 2003; Kwak and Huh, The eelgrass (Zostera marina) beds is very 2004). Worldwide, the members of family Mona- important as rich, productive nursery and feed- canthidae are associated with shallow water sea- ing area for juvenile and adult fishes (Adams, grass beds, and seagrass tissue forms major por- 1976; Pollard, 1984; Klumpp et al., 1989; Hemm- tion of their diets (e.g. -

Chinese Red Swimming Crab (Portunus Haanii) Fishery Improvement Project (FIP) in Dongshan, China (August-December 2018)
Chinese Red Swimming Crab (Portunus haanii) Fishery Improvement Project (FIP) in Dongshan, China (August-December 2018) Prepared by Min Liu & Bai-an Lin Fish Biology Laboratory College of Ocean and Earth Sciences, Xiamen University March 2019 Contents 1. Introduction........................................................................................................ 5 2. Materials and Methods ...................................................................................... 6 2.1. Study site and survey frequency .................................................................... 6 2.2. Sample collection .......................................................................................... 7 2.3. Species identification................................................................................... 10 2.4. Sample measurement ................................................................................... 11 2.5. Interviews.................................................................................................... 13 2.6. Estimation of annual capture volume of Portunus haanii ............................. 15 3. Results .............................................................................................................. 15 3.1. Species diversity.......................................................................................... 15 3.1.1. Species composition .............................................................................. 15 3.1.2. ETP species ......................................................................................... -

Marine Ecosystems of the North Pacific Ocean 2003-2008
Marine Ecosystems of the North Pacific Ocean 2003-2008 McKinnell, S.M. and Dagg, M.J. [Eds.] 2010. Marine Ecosystems of the North Pacific Ocean, 2003-2008. PICES Special Publication 4, 393 p. PICES Special Publication Number 4 Yellow and East China Seas lead author Sinjae Yoo Marine Living Resources Division Korea Ocean Research and Development Institute (KORDI) Ansan, Republic of Korea Citation: Yoo, S., An, Y.-R., Bae, S., Choi, S., Ishizaka, J., Kang, Y.-S., Kim, Z.G., Lee, C., Lee, J.B., Li, R., Park, J., Wang, Z., Wen, Q., Yang, E. J., Yeh, S.-W., Yeon, I., Yoon, W.-D., Zhang, C.-I., Zhang, X., Zhu, M. 2010. Status and trends in the Yellow Sea and East China Sea region, pp. 360-393 In S.M. McKinnell and M.J. Dagg [Eds.] Marine Ecosystems of the North Pacific Ocean, 2003-2008. PICES Special Publication 4, 393 p. Yellow and East Yellow China Seas [361] highlights The Yellow Sea and East China Sea have undergone drastic changes in the past decades as witnessed by species shifts, increasing outbreaks of HABs, and jellyfish blooms. More recently, macroalgal blooms appeared in the western coastal areas of the Yellow Sea and East China Sea (Liu et al. 2009). These and other changes indicate the ecosystem structure is rapidly shifting. There is clear evidence of ongoing eutrophication in the Yellow Sea from the record of organic deposition in sediments, increases in biomass and abundance of the benthos during the past decade, and the appearance of an hypoxic area. Warming of the ocean surface waters has occurred since the mid- to late 1980s, with a parallel increasing trend in mesozooplankton abundance. -

A Cyprinid Fish
DFO - Library / MPO - Bibliotheque 01005886 c.i FISHERIES RESEARCH BOARD OF CANADA Biological Station, Nanaimo, B.C. Circular No. 65 RUSSIAN-ENGLISH GLOSSARY OF NAMES OF AQUATIC ORGANISMS AND OTHER BIOLOGICAL AND RELATED TERMS Compiled by W. E. Ricker Fisheries Research Board of Canada Nanaimo, B.C. August, 1962 FISHERIES RESEARCH BOARD OF CANADA Biological Station, Nanaimo, B0C. Circular No. 65 9^ RUSSIAN-ENGLISH GLOSSARY OF NAMES OF AQUATIC ORGANISMS AND OTHER BIOLOGICAL AND RELATED TERMS ^5, Compiled by W. E. Ricker Fisheries Research Board of Canada Nanaimo, B.C. August, 1962 FOREWORD This short Russian-English glossary is meant to be of assistance in translating scientific articles in the fields of aquatic biology and the study of fishes and fisheries. j^ Definitions have been obtained from a variety of sources. For the names of fishes, the text volume of "Commercial Fishes of the USSR" provided English equivalents of many Russian names. Others were found in Berg's "Freshwater Fishes", and in works by Nikolsky (1954), Galkin (1958), Borisov and Ovsiannikov (1958), Martinsen (1959), and others. The kinds of fishes most emphasized are the larger species, especially those which are of importance as food fishes in the USSR, hence likely to be encountered in routine translating. However, names of a number of important commercial species in other parts of the world have been taken from Martinsen's list. For species for which no recognized English name was discovered, I have usually given either a transliteration or a translation of the Russian name; these are put in quotation marks to distinguish them from recognized English names.