What Mad Pursuit BOOKS in the ALFRED P
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Francis Crick in Molecular Biology
2019 Asia-Pacific Conference on Emerging Technologies and Engineering (ACETE 2019) Francis Crick in Molecular Biology Sun Yongping College of Physic and Electronic Information, Inner Mongolia Normal University, Hohhot, China Keywords: Crick, DNA, Protein, Genetic Codes, Molecular Biology Abstract: This article is a tribute to Francis crick, a biophysicist who passed away on July 28, 2004. Francis crick, James Watson and Maurice Wilkins were jointly awarded the 1962 Nobel Prize for physiology or medicine for discovering the molecular structure of nucleic acids and its significance for information transfer in living material. It is pointed out that the diverse background and unique sensitivity of crick to science enabled him to have great insights into frontier research. He had a special capacity for prudent and logical thinking, which contributed so much to the development of molecular biology. Based on Francis crick’s academic achievements in molecular biology and by virtue of internal history approaches such as concept analysis and literature research, this paper is aimed at revealing the historical contributions of crick in a condensed way and to commemorate his work. 1. Introduction Francis crick (figure 1) was born on June 8, 1916 as an English citizen, and he left the world, aged 88. With lifelong devotion to scientific research, crick is credited as one of the central figures in the molecular revolution that swept through biology in the latter half of the twentieth century [1]. Keen on seeking after and tackling the profound problems, he developed a passion for biology although crick did research in physics at the beginning of his scientific life [2,3]. -

Discovery of DNA Structure and Function: Watson and Crick By: Leslie A
01/08/2018 Discovery of DNA Double Helix: Watson and Crick | Learn Science at Scitable NUCLEIC ACID STRUCTURE AND FUNCTION | Lead Editor: Bob Moss Discovery of DNA Structure and Function: Watson and Crick By: Leslie A. Pray, Ph.D. © 2008 Nature Education Citation: Pray, L. (2008) Discovery of DNA structure and function: Watson and Crick. Nature Education 1(1):100 The landmark ideas of Watson and Crick relied heavily on the work of other scientists. What did the duo actually discover? Aa Aa Aa Many people believe that American biologist James Watson and English physicist Francis Crick discovered DNA in the 1950s. In reality, this is not the case. Rather, DNA was first identified in the late 1860s by Swiss chemist Friedrich Miescher. Then, in the decades following Miescher's discovery, other scientists--notably, Phoebus Levene and Erwin Chargaff--carried out a series of research efforts that revealed additional details about the DNA molecule, including its primary chemical components and the ways in which they joined with one another. Without the scientific foundation provided by these pioneers, Watson and Crick may never have reached their groundbreaking conclusion of 1953: that the DNA molecule exists in the form of a three-dimensional double helix. The First Piece of the Puzzle: Miescher Discovers DNA Although few people realize it, 1869 was a landmark year in genetic research, because it was the year in which Swiss physiological chemist Friedrich Miescher first identified what he called "nuclein" inside the nuclei of human white blood cells. (The term "nuclein" was later changed to "nucleic acid" and eventually to "deoxyribonucleic acid," or "DNA.") Miescher's plan was to isolate and characterize not the nuclein (which nobody at that time realized existed) but instead the protein components of leukocytes (white blood cells). -

Biochemistrystanford00kornrich.Pdf
University of California Berkeley Regional Oral History Office University of California The Bancroft Library Berkeley, California Program in the History of the Biosciences and Biotechnology Arthur Kornberg, M.D. BIOCHEMISTRY AT STANFORD, BIOTECHNOLOGY AT DNAX With an Introduction by Joshua Lederberg Interviews Conducted by Sally Smith Hughes, Ph.D. in 1997 Copyright 1998 by The Regents of the University of California Since 1954 the Regional Oral History Office has been interviewing leading participants in or well-placed witnesses to major events in the development of Northern California, the West, and the Nation. Oral history is a method of collecting historical information through tape-recorded interviews between a narrator with firsthand knowledge of historically significant events and a well- informed interviewer, with the goal of preserving substantive additions to the historical record. The tape recording is transcribed, lightly edited for continuity and clarity, and reviewed by the interviewee. The corrected manuscript is indexed, bound with photographs and illustrative materials, and placed in The Bancroft Library at the University of California, Berkeley, and in other research collections for scholarly use. Because it is primary material, oral history is not intended to present the final, verified, or complete narrative of events. It is a spoken account, offered by the interviewee in response to questioning, and as such it is reflective, partisan, deeply involved, and irreplaceable. ************************************ All uses of this manuscript are covered by a legal agreement between The Regents of the University of California and Arthur Kornberg, M.D., dated June 18, 1997. The manuscript is thereby made available for research purposes. All literary rights in the manuscript, including the right to publish, are reserved to The Bancroft Library of the University of California, Berkeley. -

Francis HC Crick
Francis H. C. Crick: memories of a friend of Francis and Odile The world feels strange without Francis. It is of course full of memories of him. Mostly memories of the charismatic personality, of the brilliant mind, of the great scientist, of the stories of his discoveries in molecular biology. After all, the names of Watson and Crick will be with us as long as Einstein’s and Planck’s. My fondest memories of Francis are of a different kind. I met him at about the time – in ‘76 – when Francis and Odile moved from Cambridge, England to the Salk Institute in La Jolla, California. At the Salk he became a theoretical neuroscientist, following his second passion. After the mystery of life, the mystery of the mind. I saw him at F.O. Schmitt ‘s Neuroscience Research Program meetings. I visited Francis and Odile during the summers in their house on Portugal Place in Cambridge, England, with its golden helix above the front door. I went with them and the Orgels in trips to the desert. I saw him debating about consciousness with various guests at my home. In ‘79, I worked for an intense month at the Salk Institute with him and the late David Marr, trying to understand the connection between the architecture of visual cortex and several intriguing aspects of our visual perception. In those years molecular biology was becoming the dominant science. I remember the difficulty – then, not now -- of getting neuroscience appointments through the well- earned intellectual arrogance of our friends and colleagues in the Department of Biology at MIT. -
BSCB Newsletter 2017D
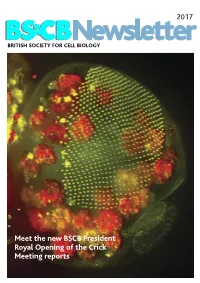
2017 BSCB Newsletter BRITISH SOCIETY FOR CELL BIOLOGY Meet the new BSCB President Royal Opening of the Crick Meeting reports 2017 CONTENTS BSCB Newsletter News 2 Book reviews 7 Features 8 Meeting Reports 24 Summer students 30 Society Business 33 Editorial Welcome to the 2017 BSCB newsletter. After several meeting hosted several well received events for our Front cover: years of excellent service, Kate Nobes has stepped PhD and Postdoc members, which we discuss on The head of a Drosophila pupa. The developing down and handed the reins over to me. I’ve enjoyed page 5. Our PhD and Postdoc reps are working hard compound eye (green) is putting together this years’ newsletter. It’s been great to make the event bigger and better for next year! The composed of several hundred simple units called ommatidia to hear what our members have been up to, and I social events were well attended including the now arranged in an extremely hope you will enjoy reading it. infamous annual “Pub Quiz” and disco after the regular array. The giant conference dinner. Members will be relieved to know polyploidy cells of the fat body (red), the fly equivalent of the The 2016 BSCB/DB spring meeting, organised by our we aren’t including any photos from that here. mammalian liver and adipose committee members Buzz Baum (UCL), Silke tissue, occupy a big area of the Robatzek and Steve Royle, had a particular focus on In this issue, we highlight the great work the BSCB head. Cells and Tissue Architecture, Growth & Cell Division, has been doing to engage young scientists. -

Socity the Physiologicalsociety Newsletter
p rVI i~ ne Pal Newsette Socity The PhysiologicalSociety Newsletter Contents 1 Physiological Sciences at Oxford - Clive Ellory 2 Neuroscience Research at Monash University - Uwe Proske 4 Committee News 4 Grants for IUPS Congress, Glasgow, 1993 4 COPUS - Committee on the Public Understanding of Science 4 Nominations for election to Membership 5 Computers in Teaching Initiative 5 Membership Subscriptions for 1993 5 Benevolent Fund 6 Wellcome Prize Lecturer 6 Retiring Committee Members 8 Letters & Reports 8 Society's Meetings 9 Animal Research - Speaking in Schools 9 Colin Blakemore - FRS 9 Happy 80th Birthday 10 Talking Point in the Biological Sciences - Simon Brophy,RDS 11 Chance & Design 11 Views 11 Muscular Dystrophy Group - SarahYates 14 The Multiple Sclerosis Society - John Walford 14 British Diabetic Association - Moira Murphy 15 Biomedical Research in the SERC - Alan Thomas 16 Cancer Research Campaign - TA Hince 18 The Wellcome Trust - JulianJack 22 Articles 22 Immunosuppression in Multiple Sclerosis - A N Davison 23 Hypoxia - a regulator of uterine contractions in labour? - Susan Wray 25 Pregnancy and the vascular endothelium - Lucilla Poston 28 Society Sponsored Events 28 IUPS Congress 93, list of themes 32 Notices 35 Tear-Out Forms 35 Affiliates 37 Grey Book Updates Administrations & Publications Office, P 0 Box 506, Oxford, OXI 3XE Tel: (0865) 798498 Fax: (0865) 798092 Produced by Kwabena Appenteng, Heather Dalitz and Clare Haigh The PhysiofogicafSociety 9ewsfetter Physiological Sciences at Oxford The two year interval since the last meeting of the Society in Lecturer in the department for some time, has been appointed to Oxford corresponds with the time I have been standing in for a university lectureship, in association with Balliol College. -

Network AW 2004.Qxd 10/11/2004 13:40 Page 1
Network AW 2004.qxd 10/11/2004 13:40 Page 1 network Autumn/Winter 2004 News, views and information from the Medical Research Council In this issue... MRC integral to UK clinical Funding update research drive News of competition for MRC funding in Speaking at the Academy of Medical Sciences (AMS) in 2004/05 and the application June, Lord Warner of Brockley announced the formation review process of the UK Clinical Research Collaboration (UKCRC) – a page 2 multi-partner initiative launched to give a major boost to clinical research in the UK.The Government has also set up a new MRC/Health Departments (HDs) Joint Health Francis Crick Delivery Group to increase the strategic coordination of 1916-2004 publicly funded medical research and support the UKCRC. A colleague pays tribute to one of the founding fathers UKCRC of molecular biology The UKCRC was a key recommendation of the Research page 4 for Patient Benefit Working Party set up in response to "We welcome Chancellor Gordon Brown's budget influential reports in Autumn 2003 by the AMS and the announcement of a cash injection for medical research Government's Bioscience Innovation and Growth Team. over the next four years, and will be working even more Basic Technology With a mission to coordinate and transform clinical closely with the Government and the Health Departments November showcase for research in the UK, the partnership will involve the MRC, to address national health priorities in the years ahead." progress by MRC scientists Government, the NHS, academia, medical charities, Colin Blakemore, MRC Chief Executive in the Research Councils UK industry and the public. -

The Use of Non-Human Primates in Research in Primates Non-Human of Use The
The use of non-human primates in research The use of non-human primates in research A working group report chaired by Sir David Weatherall FRS FMedSci Report sponsored by: Academy of Medical Sciences Medical Research Council The Royal Society Wellcome Trust 10 Carlton House Terrace 20 Park Crescent 6-9 Carlton House Terrace 215 Euston Road London, SW1Y 5AH London, W1B 1AL London, SW1Y 5AG London, NW1 2BE December 2006 December Tel: +44(0)20 7969 5288 Tel: +44(0)20 7636 5422 Tel: +44(0)20 7451 2590 Tel: +44(0)20 7611 8888 Fax: +44(0)20 7969 5298 Fax: +44(0)20 7436 6179 Fax: +44(0)20 7451 2692 Fax: +44(0)20 7611 8545 Email: E-mail: E-mail: E-mail: [email protected] [email protected] [email protected] [email protected] Web: www.acmedsci.ac.uk Web: www.mrc.ac.uk Web: www.royalsoc.ac.uk Web: www.wellcome.ac.uk December 2006 The use of non-human primates in research A working group report chaired by Sir David Weatheall FRS FMedSci December 2006 Sponsors’ statement The use of non-human primates continues to be one the most contentious areas of biological and medical research. The publication of this independent report into the scientific basis for the past, current and future role of non-human primates in research is both a necessary and timely contribution to the debate. We emphasise that members of the working group have worked independently of the four sponsoring organisations. Our organisations did not provide input into the report’s content, conclusions or recommendations. -

Science in Action
SCIENCE IN ACTION How to follow scientists and engineers through society Bruno Latour Harvard UnlvetSHy Press Cambridge, Massachusetts 1987 INTRODUCTION Opening Pandora's Black Box Scene 1: On a cold and sunny morning in October 1985, John Whittaker entered his office in the molecular biology building of the Institut Pasteur in Paris and switched on his Eclipse MV/8000 computer. A few seconds after loading the special programs he had written, a three-dimensional picture of the DNA double helix flashed onto the screen. John, a visiting computer scientist, had been invited by the Institute to write programs that could produce three-dimensional images of the coils of DNA and relate them to the thousands of new nucleic acid sequences pouring out every year into the journals and data banks. 'Nice picture, eh?' said his boss, Pierre, who was just entering the office. 'Yes, good machine too,' answered John. Scene 2: In 1951 in the Cavendish laboratory at Cambridge, England, the X-ray pictures of crystallised deoxyribonucleic acid were not 'nice pictures' on a computer screen. The two young researchers, Jim Watson and Francis Crick1, had a hard time obtaining them from Maurice Wilkins and Rosalind Franklin in London. It was impossible yet to decide if the form of the acid was a triple or a double helix, if the phosphate bonds were at the inside or at the outside of the molecule, or indeed if it was an helix at all. It did not matter much to their boss, Sir Francis Bragg, since the two were not supposed to be working on DNA anyway, but it mattered a lot to them, especially since Linus Pauling, the famous chemist, was said to be about to uncover the structure of DNA in a few months. -

Annual Report Fy 2016
PHILLIPS BROOKS HOUSE ASSOCIATION “E ve ANNUAL REPORT FY 2016 phillips brooks house association “Each time a man stands up for an ideal, or acts to improve the lot of others, or strikes out against injustice, he sends forth a tiny ripple of hope, and those ripples build a current which can sweep down the mightiest walls of oppression and resistance.” -Robert F. Kennedy 2 | PBHA ANNUAL REPORT Dear PBHA Supporters, Phillips Brooks House Association’s 2015 was a truly remark- able year and one that illustrates, perhaps more than ever, the power and impact of what we can accomplish together. This year we were so proud to support the creation and opening of Y2Y (Youth to Youth) Harvard Square, a youth shelter which, thanks to the leadership of alumni Sam Green- berg and Sarah Rosenkrantz, is a powerful example of how we can address some of society’s greatest needs by building partnerships. Y2Y’s opening, which followed an extensive renovation of the space located at First Parish in Cambridge, Unitarian Universalist, united students, homeless youth, residents, business owners, elected officials, and donors in the shared mission of tripling the number of shelter beds dedicated to 18-24 year-olds in Greater Boston. HOPE, the Harvard Organization for Prison Education and Reform, built connections between the prison education programs that have been part of PBHA for more than 60 years and strengthened advocacy efforts addressing abuses in the criminal justice system. With the help of the 2015 Robert Coles Call of Service lecturer and honoree, Black Lives Matter co-founder Alicia Garza, Boston and Cambridge youth joined with Harvard student groups to show their commitment to the ideals of this critical movement. -

DNA: the Timeline and Evidence of Discovery
1/19/2017 DNA: The Timeline and Evidence of Discovery Interactive Click and Learn (Ann Brokaw Rocky River High School) Introduction For almost a century, many scientists paved the way to the ultimate discovery of DNA and its double helix structure. Without the work of these pioneering scientists, Watson and Crick may never have made their ground-breaking double helix model, published in 1953. The knowledge of how genetic material is stored and copied in this molecule gave rise to a new way of looking at and manipulating biological processes, called molecular biology. The breakthrough changed the face of biology and our lives forever. Watch The Double Helix short film (approximately 15 minutes) – hyperlinked here. 1 1/19/2017 1865 The Garden Pea 1865 The Garden Pea In 1865, Gregor Mendel established the foundation of genetics by unraveling the basic principles of heredity, though his work would not be recognized as “revolutionary” until after his death. By studying the common garden pea plant, Mendel demonstrated the inheritance of “discrete units” and introduced the idea that the inheritance of these units from generation to generation follows particular patterns. These patterns are now referred to as the “Laws of Mendelian Inheritance.” 2 1/19/2017 1869 The Isolation of “Nuclein” 1869 Isolated Nuclein Friedrich Miescher, a Swiss researcher, noticed an unknown precipitate in his work with white blood cells. Upon isolating the material, he noted that it resisted protein-digesting enzymes. Why is it important that the material was not digested by the enzymes? Further work led him to the discovery that the substance contained carbon, hydrogen, nitrogen and large amounts of phosphorus with no sulfur. -

The Gene in the Twenty-First Century
CGA_C01.qxd 4/24/07 10:16 Page 1 1 CHAPTER 1 The gene in the twenty-first century Choong-Chin Liew, PhD, & Victor J. Dzau, MD Vries named the transmitted substances “pangens”; Introduction he later coined the term “mutation” to signify the When the word was first used in 1909, “gene” was a appearance of a new pangen [5]. hypothesis necessary to explain puzzling observa- Cambridge evolutionist William Bateson (1861– tions about heredity. As the century progressed, the 1926) translated Mendel into English and worked hypothesis began to acquire reality as the structure vigourously to promote Mendel’s ideas in the and functions of the gene were gradually elucidated. English-speaking scientific world. Bateson himself Earlier and simpler concepts became superseded as coined the term “genetics” in 1906 [6]. The word evidence led to better understanding of the gene. “gene” was not introduced until 1909, when Wilhelm Today the gene is recognized to be a highly complex Johannsen (1857–1927), a Danish botanist, offered entity. The genomics revolution is well underway this term in preference to earlier terms [7]. but there is much that remains for twenty-first cen- A next major step towards an elucidation of tury science to learn before the potential of molec- the gene came with the discovery that genes have ular biology and technology can be fully realized. physical locations on chromosomes in studies on Drosophila carried out by Thomas Hunt Morgan (1866–1945) and his colleagues at the zoology de- The search for the gene partment of Columbia University [8,9]. Morgan’s Much of the science that laid the foundation for the student, Alfred Sturtevant (1891–1970) was able to genetics and genomics revolution took place in the show that the gene for a trait was localized in a fixed very near past; 1900 is the date often considered to location or locus arranged “like beads on a string” be the beginning of modern genetics.