The Metacyc Database of Metabolic Pathways and Enzymes - a 2019 Update Ron Caspi*, Richard Billington, Ingrid M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Diversity of Understudied Archaeal and Bacterial Populations of Yellowstone National Park: from Genes to Genomes Daniel Colman
University of New Mexico UNM Digital Repository Biology ETDs Electronic Theses and Dissertations 7-1-2015 Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes Daniel Colman Follow this and additional works at: https://digitalrepository.unm.edu/biol_etds Recommended Citation Colman, Daniel. "Diversity of understudied archaeal and bacterial populations of Yellowstone National Park: from genes to genomes." (2015). https://digitalrepository.unm.edu/biol_etds/18 This Dissertation is brought to you for free and open access by the Electronic Theses and Dissertations at UNM Digital Repository. It has been accepted for inclusion in Biology ETDs by an authorized administrator of UNM Digital Repository. For more information, please contact [email protected]. Daniel Robert Colman Candidate Biology Department This dissertation is approved, and it is acceptable in quality and form for publication: Approved by the Dissertation Committee: Cristina Takacs-Vesbach , Chairperson Robert Sinsabaugh Laura Crossey Diana Northup i Diversity of understudied archaeal and bacterial populations from Yellowstone National Park: from genes to genomes by Daniel Robert Colman B.S. Biology, University of New Mexico, 2009 DISSERTATION Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Biology The University of New Mexico Albuquerque, New Mexico July 2015 ii DEDICATION I would like to dedicate this dissertation to my late grandfather, Kenneth Leo Colman, associate professor of Animal Science in the Wool laboratory at Montana State University, who even very near the end of his earthly tenure, thought it pertinent to quiz my knowledge of oxidized nitrogen compounds. He was a man of great curiosity about the natural world, and to whom I owe an acknowledgement for his legacy of intellectual (and actual) wanderlust. -

Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline
Phylogenetics of Archaeal Lipids Amy Kelly 9/27/2006 Outline • Phlogenetics of Archaea • Phlogenetics of archaeal lipids • Papers Phyla • Two? main phyla – Euryarchaeota • Methanogens • Extreme halophiles • Extreme thermophiles • Sulfate-reducing – Crenarchaeota • Extreme thermophiles – Korarchaeota? • Hyperthermophiles • indicated only by environmental DNA sequences – Nanoarchaeum? • N. equitans a fast evolving euryarchaeal lineage, not novel, early diverging archaeal phylum – Ancient archael group? • In deepest brances of Crenarchaea? Euryarchaea? Archaeal Lipids • Methanogens – Di- and tetra-ethers of glycerol and isoprenoid alcohols – Core mostly archaeol or caldarchaeol – Core sometimes sn-2- or Images removed due to sn-3-hydroxyarchaeol or copyright considerations. macrocyclic archaeol –PMI • Halophiles – Similar to methanogens – Exclusively synthesize bacterioruberin • Marine Crenarchaea Depositional Archaeal Lipids Biological Origin Environment Crocetane methanotrophs? methane seeps? methanogens, PMI (2,6,10,15,19-pentamethylicosane) methanotrophs hypersaline, anoxic Squalane hypersaline? C31-C40 head-to-head isoprenoids Smit & Mushegian • “Lost” enzymes of MVA pathway must exist – Phosphomevalonate kinase (PMK) – Diphosphomevalonate decarboxylase – Isopentenyl diphosphate isomerase (IPPI) Kaneda et al. 2001 Rohdich et al. 2001 Boucher et al. • Isoprenoid biosynthesis of archaea evolved through a combination of processes – Co-option of ancestral enzymes – Modification of enzymatic specificity – Orthologous and non-orthologous gene -

Modular Logic in Enzymatic Biogenesis of Yersiniabactin by Ye&N& Pestis
Research Paper 573 Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Ye&n& pestis Amy M Gehring l* , Edward DeMoll*, Jacqueline D Fetherston*, lchiro Moril+, George F Mayhew3, Frederick R Blattner3, Christopher T Walsh’ and Robert D Perry* Background: Virulence in the pathogenic bacterium Yersinia pestis, causative Addresses: 1 Department of Biological Chemistry agent of bubonic plague, has been correlated with the biosynthesis and and Molecular Pharmacology, Harvard Medical School, 240 Longwood Avenue, Boston, MA transport of an iron-chelating siderophore, yersiniabactin, which is induced 02115, USA. *Department of Microbiology and under iron-starvation conditions. Initial DNA sequencing suggested that this Immunology, MS41 5 Medical Center, University of system is highly conserved among the pathogenic Yersinia. Yersiniabactin Kentucky, Lexington, KY 40536-0084, USA. contains a phenolic group and three five-membered thiazole heterocycles that 3Department of Genetics, University of Wisconsin, 445 Henry Hall, Madison, WI 53706, USA. serve as iron ligands. Present addresses: ‘Department of Microbiology Results: The entire Y. pestis yersiniabactin region has been sequenced. and Cellular Biology, Harvard University, Sequence analysis of yersiniabactin biosynthetic regions (irp2-ybtf and ybtS) Cambridge, MA 02115, USA. +Nippon Glaxo Ltd. Tsukuba Research Laboratories, Chemistry reveals a strategy for siderophore production using a mixed polyketide synthase/ Department, Ibaraki, Japan. nonribosomal peptide synthetase complex formed between HMWPl and HMWPS (encoded by irpl and irp2). The complex contains 16 domains, five of Correspondence: Christopher T Walsh them variants of phosphopantetheine-modified peptidyl carrier protein or acyl E-mail: [email protected] carrier protein domains. HMWPl and HMWPP also contain methyltransferase Key words: heterocyclization, nonribosomal and heterocyclization domains. -

Chemical and Biological Aspects of Nutritional Immunity
This is a repository copy of Chemical and Biological Aspects of Nutritional Immunity - Perspectives for New Anti-infectives Targeting Iron Uptake Systems : Perspectives for New Anti-infectives Targeting Iron Uptake Systems. White Rose Research Online URL for this paper: https://eprints.whiterose.ac.uk/119363/ Version: Accepted Version Article: Bilitewski, Ursula, Blodgett, Joshua A.V., Duhme-Klair, Anne Kathrin orcid.org/0000-0001- 6214-2459 et al. (4 more authors) (2017) Chemical and Biological Aspects of Nutritional Immunity - Perspectives for New Anti-infectives Targeting Iron Uptake Systems : Perspectives for New Anti-infectives Targeting Iron Uptake Systems. Angewandte Chemie International Edition. pp. 2-25. ISSN 1433-7851 https://doi.org/10.1002/anie.201701586 Reuse Items deposited in White Rose Research Online are protected by copyright, with all rights reserved unless indicated otherwise. They may be downloaded and/or printed for private study, or other acts as permitted by national copyright laws. The publisher or other rights holders may allow further reproduction and re-use of the full text version. This is indicated by the licence information on the White Rose Research Online record for the item. Takedown If you consider content in White Rose Research Online to be in breach of UK law, please notify us by emailing [email protected] including the URL of the record and the reason for the withdrawal request. [email protected] https://eprints.whiterose.ac.uk/ AngewandteA Journal of the Gesellschaft Deutscher Chemiker International Edition Chemie www.angewandte.org Accepted Article Title: Chemical and Biological Aspects of Nutritional Immunity - Perspectives for New Anti-infectives Targeting Iron Uptake Systems Authors: Sabine Laschat, Ursula Bilitewski, Joshua Blodgett, Anne- Kathrin Duhme-Klair, Sabrina Dallavalle, Anne Routledge, and Rainer Schobert This manuscript has been accepted after peer review and appears as an Accepted Article online prior to editing, proofing, and formal publication of the final Version of Record (VoR). -
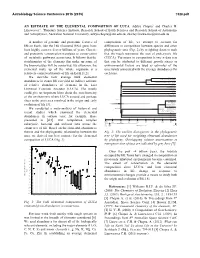
An Estimate of the Elemental Composition of Luca
Astrobiology Science Conference 2015 (2015) 7328.pdf AN ESTIMATE OF THE ELEMENTAL COMPOSITION OF LUCA. Aditya Chopra1 and Charles H. Lineweaver1, 1Planetary Science Institute, Research School of Earth Sciences and Research School of Astronomy and Astrophysics, Australian National University, [email protected], [email protected] A number of genomic and proteomic features of composition of life, we attempt to account for life on Earth, like the 16S ribosomal RNA gene, have differences in composition between species and other been highly conserved over billions of years. Genetic phylogenetic taxa (Fig. 2) by weighting datasets such and proteomic conservation translates to conservation that the result represents the root of prokaryotic life of metabolic pathways across taxa. It follows that the (LUCA). Variations in composition between data sets stoichiometry of the elements that make up some of that can be attributed to different growth stages or the biomolecules will be conserved. By extension, the environmental factors are used as estimates of the elemental make up of the whole organism is a uncertainty associated with the average abundances for relatively conserved feature of life on Earth [1,2]. each taxa. We describe how average bulk elemental Euryarchaeota 1a Methanococci, Methanobacteria, Methanopyri 7 Euryarchaeota 1b abundances in extant life can yield an indirect estimate 6 Thermoplasmata, Methanomicrobia, Halobacteria, Archaeoglobi Euryarchaeota 2 4 Thermococci of relative abundances of elements in the Last Crenarchaeota Sulfolobus, Thermoproteus ? Thaumarchaeota Universal Common Ancestor (LUCA). The results Cenarchaeum ? Korarchaeota ARMAN could give us important hints about the stoichiometry ? 2 Archaeal Richmond Mine Acidophilic Nanoorganisms Nanoarchaeota of the environment where LUCA existed and perhaps Archaea Terrabacteria clues to the processes involved in the origin and early Actinobacteria, Deinococcus-Thermus, 1 Cyanobacteria, Life Chloroflexi, 9 evolution of life [3]. -

Nuevos Avances En La RMN Anisotrópica Y Detección De Productos Naturales Marinos Bioactivos
DEPARTAMENTO DE QUÍMICA FUNDAMENTAL Programa regulado por el RD 1393/2007: Química Ambiental y Fundamental New Advances on Anisotropic NMR and Detection of Bioactive Marine Natural Products Nuevos Avances en la RMN Anisotrópica y Detección de Productos Naturales Marinos Bioactivos Memoria presentada por Juan Carlos C. Fuente Monteverde Directores: Dr. Jaime Rodríguez González Dr. Carlos Jiménez González A Coruña 2018 i ii Acta de Tesis El tribunal, nombrado por el Excmo. Sr. Rector de la Universidade da Coruña para calificar la tesis doctoral titulada “Nuevos Avances en la RMN Anisotrópica y Detección de Productos Naturales Marinos Bioactivos/New Advances on Anisotropic NMR and Detection of Bioactive Marine Natural Products” dirigida por los Drs. Jaime Rodríguez González y Carlos Jiménez González, presentada por Don Juan Carlos de la Cruz Fuentes Monteverde y constituido en el día de la fecha por los miembros que subscriben la presente Acta, una vez efectuada la defensa por el doctorando y contestadas las objeciones y/o sugerencias que se le han formulado, ha otorgado por …………………………la calificación de: En A Coruña, a……………… de ……………………………… de 2019 PRESIDENTE SECRETARIO VOCAL Prof. Dr. Michael Reggelin Pr. Dr. Marcos D. García Romero Pr. Dr. Victor Sanchez Pedregal Organic Chemistry Pr. Titular Química Orgánica Pr. Titular de Química Orgánica Technische Universität Darmstadt Firmado Firmado Firmado iii iv Don Juan Carlos de la Cruz Fuentes Monteverde: Presenta la memoria adjunta, titulada “Nuevos Avances en la RMN Anisotrópica y Detección de Productos Naturales Marinos Bioactivos/New Advances on Anisotropic NMR and Detection of Bioactive Marine Natural Products” para optar al grado de Doctor en Química que ha sido realizado bajo la dirección de los Doctores Jaime Rodríguez González y Carlos Jiménez González en los laboratorios del Centro de Investigaciones Científicas Avanzadas (CICA) de la Universidade da Coruña. -

Interrupted Adenylation Domains: Unique Bifunctional Enzymes Involved in Nonribosomal Peptide Biosynthesis
Natural Product Reports Interrupted adenylation domains: unique bifunctional enzymes involved in nonribosomal peptide biosynthesis Journal: Natural Product Reports Manuscript ID: NP-REV-09-2014-000120.R1 Article Type: Highlight Date Submitted by the Author: 12-Jan-2015 Complete List of Authors: Labby, Kristin; Beloit College, Chemistry Watsula, Stoyan; University of Michigan, Medicinal Chemistry Garneau-Tsodikova, Sylvie; University of Kentucky, Pharmaceutical Sciences Page 1 of 11NPR Natural Product Reports Dynamic Article Links ► Cite this: DOI: 10.1039/c0xx00000x www.rsc.org/xxxxxx HIGHLIGHT Interrupted adenylation domains: unique bifunctional enzymes involved in nonribosomal peptide biosynthesis Kristin J. Labby, a Stoyan G. Watsula,b and Sylvie Garneau-Tsodikova* c Received (in XXX, XXX) Xth XXXXXXXXX 20XX, Accepted Xth XXXXXXXXX 20XX 5 DOI: 10.1039/b000000x Covering up to 2014 Nonribosomal peptides (NRPs) account for a large portion of drugs and drug leads currently available in the pharmaceutical industry. They are one of two main families of natural products biosynthesized on megaenzyme assembly-lines composed of multiple modules that are, in general, each comprised of three 10 core domains and on occasion of accompanying auxiliary domains. The core adenylation (A) domains are known to delineate the identity of the specific chemical components to be incorporated into the growing NRPs. Previously believed to be inactive, A domains interrupted by auxiliary enzymes have recently been proven to be active and capable of performing two distinct chemical reactions. This highlight summarizes current knowledge on A domains and presents the various interrupted A domains found in a number of 15 nonribosomal peptide synthetase (NRPS) assembly-lines, their predicted or proven dual functions, and their potential for manipulation and engineering for chemoenzymatic synthesis of new pharmaceutical agents with increased potency. -

Pan-Genome Analysis and Ancestral State Reconstruction Of
www.nature.com/scientificreports OPEN Pan‑genome analysis and ancestral state reconstruction of class halobacteria: probability of a new super‑order Sonam Gaba1,2, Abha Kumari2, Marnix Medema 3 & Rajeev Kaushik1* Halobacteria, a class of Euryarchaeota are extremely halophilic archaea that can adapt to a wide range of salt concentration generally from 10% NaCl to saturated salt concentration of 32% NaCl. It consists of the orders: Halobacteriales, Haloferaciales and Natriabales. Pan‑genome analysis of class Halobacteria was done to explore the core (300) and variable components (Softcore: 998, Cloud:36531, Shell:11784). The core component revealed genes of replication, transcription, translation and repair, whereas the variable component had a major portion of environmental information processing. The pan‑gene matrix was mapped onto the core‑gene tree to fnd the ancestral (44.8%) and derived genes (55.1%) of the Last Common Ancestor of Halobacteria. A High percentage of derived genes along with presence of transformation and conjugation genes indicate the occurrence of horizontal gene transfer during the evolution of Halobacteria. A Core and pan‑gene tree were also constructed to infer a phylogeny which implicated on the new super‑order comprising of Natrialbales and Halobacteriales. Halobacteria1,2 is a class of phylum Euryarchaeota3 consisting of extremely halophilic archaea found till date and contains three orders namely Halobacteriales4,5 Haloferacales5 and Natrialbales5. Tese microorganisms are able to dwell at wide range of salt concentration generally from 10% NaCl to saturated salt concentration of 32% NaCl6. Halobacteria, as the name suggests were once considered a part of a domain "Bacteria" but with the discovery of the third domain "Archaea" by Carl Woese et al.7, it became part of Archaea. -

Variations in the Two Last Steps of the Purine Biosynthetic Pathway in Prokaryotes
GBE Different Ways of Doing the Same: Variations in the Two Last Steps of the Purine Biosynthetic Pathway in Prokaryotes Dennifier Costa Brandao~ Cruz1, Lenon Lima Santana1, Alexandre Siqueira Guedes2, Jorge Teodoro de Souza3,*, and Phellippe Arthur Santos Marbach1,* 1CCAAB, Biological Sciences, Recoˆ ncavo da Bahia Federal University, Cruz das Almas, Bahia, Brazil 2Agronomy School, Federal University of Goias, Goiania,^ Goias, Brazil 3 Department of Phytopathology, Federal University of Lavras, Minas Gerais, Brazil Downloaded from https://academic.oup.com/gbe/article/11/4/1235/5345563 by guest on 27 September 2021 *Corresponding authors: E-mails: [email protected]fla.br; [email protected]. Accepted: February 16, 2019 Abstract The last two steps of the purine biosynthetic pathway may be catalyzed by different enzymes in prokaryotes. The genes that encode these enzymes include homologs of purH, purP, purO and those encoding the AICARFT and IMPCH domains of PurH, here named purV and purJ, respectively. In Bacteria, these reactions are mainly catalyzed by the domains AICARFT and IMPCH of PurH. In Archaea, these reactions may be carried out by PurH and also by PurP and PurO, both considered signatures of this domain and analogous to the AICARFT and IMPCH domains of PurH, respectively. These genes were searched for in 1,403 completely sequenced prokaryotic genomes publicly available. Our analyses revealed taxonomic patterns for the distribution of these genes and anticorrelations in their occurrence. The analyses of bacterial genomes revealed the existence of genes coding for PurV, PurJ, and PurO, which may no longer be considered signatures of the domain Archaea. Although highly divergent, the PurOs of Archaea and Bacteria show a high level of conservation in the amino acids of the active sites of the protein, allowing us to infer that these enzymes are analogs. -

Systema Naturae. the Classification of Living Organisms
Systema Naturae. The classification of living organisms. c Alexey B. Shipunov v. 5.601 (June 26, 2007) Preface Most of researches agree that kingdom-level classification of living things needs the special rules and principles. Two approaches are possible: (a) tree- based, Hennigian approach will look for main dichotomies inside so-called “Tree of Life”; and (b) space-based, Linnaean approach will look for the key differences inside “Natural System” multidimensional “cloud”. Despite of clear advantages of tree-like approach (easy to develop rules and algorithms; trees are self-explaining), in many cases the space-based approach is still prefer- able, because it let us to summarize any kinds of taxonomically related da- ta and to compare different classifications quite easily. This approach also lead us to four-kingdom classification, but with different groups: Monera, Protista, Vegetabilia and Animalia, which represent different steps of in- creased complexity of living things, from simple prokaryotic cell to compound Nature Precedings : doi:10.1038/npre.2007.241.2 Posted 16 Aug 2007 eukaryotic cell and further to tissue/organ cell systems. The classification Only recent taxa. Viruses are not included. Abbreviations: incertae sedis (i.s.); pro parte (p.p.); sensu lato (s.l.); sedis mutabilis (sed.m.); sedis possi- bilis (sed.poss.); sensu stricto (s.str.); status mutabilis (stat.m.); quotes for “environmental” groups; asterisk for paraphyletic* taxa. 1 Regnum Monera Superphylum Archebacteria Phylum 1. Archebacteria Classis 1(1). Euryarcheota 1 2(2). Nanoarchaeota 3(3). Crenarchaeota 2 Superphylum Bacteria 3 Phylum 2. Firmicutes 4 Classis 1(4). Thermotogae sed.m. 2(5). -

Genomic Investigation of Salmonella Isolates Recovered from a Pig Slaughtering Process in Hangzhou, China
fmicb-12-704636 July 3, 2021 Time: 17:17 # 1 ORIGINAL RESEARCH published: 08 July 2021 doi: 10.3389/fmicb.2021.704636 Genomic Investigation of Salmonella Isolates Recovered From a Pig Slaughtering Process in Hangzhou, China Beibei Wu1†, Abdelaziz Ed-Dra2†, Hang Pan3†, Chenghang Dong3, Chenghao Jia3 and Min Yue2,3,4,5* 1 Zhejiang Provincial Center for Disease Control and Prevention, Hangzhou, China, 2 Hainan Institute of Zhejiang University, Sanya, China, 3 Department of Veterinary Medicine, Institute of Preventive Veterinary Sciences, Zhejiang University College of Animal Sciences, Hangzhou, China, 4 State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, National Medical Center for Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China, 5 Zhejiang Provincial Key Laboratory of Preventive Veterinary Medicine, Hangzhou, China The pig industry is the principal source of meat products in China, and the presence Edited by: of pathogens in pig-borne meat is a crucial threat to public health. Salmonella is the Bojana Bogovic Matijasic, major pathogen associated with pig-borne diseases. However, route surveillance by University of Ljubljana, Slovenia genomic platforms along the food chain is still limited in China. Here, we conducted Reviewed by: Baowei Yang, a study to evaluate the dynamic prevalence of Salmonella in a pig slaughtering Northwest A&F University, China process in Hangzhou, Zhejiang Province, China. Fifty-five of 226 (24.37%) samples Jing Han, were positive for Salmonella; from them, 78 different isolates were selected and National Center for Toxicological Research (FDA), United States subjected to whole genome sequencing followed by bioinformatics analyses to *Correspondence: determine serovar distribution, MLST patterns, antimicrobial resistance genes, plasmid Min Yue replicons, and virulence factors. -

Yersiniabactin Siderophore of Crohn's Disease-Associated Adherent
International Journal of Molecular Sciences Article Yersiniabactin Siderophore of Crohn’s Disease-Associated Adherent-Invasive Escherichia coli Is Involved in Autophagy Activation in Host Cells Guillaume Dalmasso 1,*,† , Hang Thi Thu Nguyen 1,† , Tiphanie Faïs 1,2,Sébastien Massier 1, Caroline Chevarin 1, Emilie Vazeille 1,3, Nicolas Barnich 1 , Julien Delmas 1,2 and Richard Bonnet 1,2,4,* 1 M2iSH (Microbes, Intestin, Inflammation and Susceptibility of the Host), Inserm U1071, INRAE USC 2018, Université Clermont Auvergne, CRNH, 63001 Clermont-Ferrand, France; [email protected] (H.T.T.N.); [email protected] (T.F.); [email protected] (S.M.); [email protected] (C.C.); [email protected] (E.V.); [email protected] (N.B.); [email protected] (J.D.) 2 Laboratoire de Bactériologie, Centre Hospitalier Universitaire, 63001 Clermont-Ferrand, France 3 Service d’Hépato-Gastro Entérologie, 3iHP, Centre Hospitalier Universitaire, 63000 Clermont-Ferrand, France 4 Centre de Référence de la Résistance Aux Antibiotiques, Centre Hospitalier Universitaire, 63000 Clermont-Ferrand, France * Correspondence: [email protected] (G.D.); [email protected] (R.B.) † These authors contributed equally. Abstract: Background: Adherent-invasive Escherichia coli (AIEC) have been implicated in the etiology of Crohn’s disease. The AIEC reference strain LF82 possesses a pathogenicity island similar to the high pathogenicity island of Yersinia spp., which encodes the yersiniabactin siderophore required for Citation: Dalmasso, G.; Nguyen, iron uptake and growth of the bacteria in iron-restricted environment. Here, we investigated the role H.T.T.; Faïs, T.; Massier, S.; Chevarin, of yersiniabactin during AIEC infection.