Comprehensive Evaluation of Genetic Population Structure for Anadromous T River Herring with Single Nucleotide Polymorphism Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Penobscot Rivershed with Licensed Dischargers and Critical Salmon
0# North West Branch St John T11 R15 WELS T11 R17 WELS T11 R16 WELS T11 R14 WELS T11 R13 WELS T11 R12 WELS T11 R11 WELS T11 R10 WELS T11 R9 WELS T11 R8 WELS Aroostook River Oxbow Smith Farm DamXW St John River T11 R7 WELS Garfield Plt T11 R4 WELS Chapman Ashland Machias River Stream Carry Brook Chemquasabamticook Stream Squa Pan Stream XW Daaquam River XW Whitney Bk Dam Mars Hill Squa Pan Dam Burntland Stream DamXW Westfield Prestile Stream Presque Isle Stream FRESH WAY, INC Allagash River South Branch Machias River Big Ten Twp T10 R16 WELS T10 R15 WELS T10 R14 WELS T10 R13 WELS T10 R12 WELS T10 R11 WELS T10 R10 WELS T10 R9 WELS T10 R8 WELS 0# MARS HILL UTILITY DISTRICT T10 R3 WELS Water District Resevoir Dam T10 R7 WELS T10 R6 WELS Masardis Squapan Twp XW Mars Hill DamXW Mule Brook Penobscot RiverYosungs Lakeh DamXWed0# Southwest Branch St John Blackwater River West Branch Presque Isle Strea Allagash River North Branch Blackwater River East Branch Presque Isle Strea Blaine Churchill Lake DamXW Southwest Branch St John E Twp XW Robinson Dam Prestile Stream S Otter Brook L Saint Croix Stream Cox Patent E with Licensed Dischargers and W Snare Brook T9 R8 WELS 8 T9 R17 WELS T9 R16 WELS T9 R15 WELS T9 R14 WELS 1 T9 R12 WELS T9 R11 WELS T9 R10 WELS T9 R9 WELS Mooseleuk Stream Oxbow Plt R T9 R13 WELS Houlton Brook T9 R7 WELS Aroostook River T9 R4 WELS T9 R3 WELS 9 Chandler Stream Bridgewater T T9 R5 WELS TD R2 WELS Baker Branch Critical UmScolcus Stream lmon Habitat Overlay South Branch Russell Brook Aikens Brook West Branch Umcolcus Steam LaPomkeag Stream West Branch Umcolcus Stream Tie Camp Brook Soper Brook Beaver Brook Munsungan Stream S L T8 R18 WELS T8 R17 WELS T8 R16 WELS T8 R15 WELS T8 R14 WELS Eagle Lake Twp T8 R10 WELS East Branch Howe Brook E Soper Mountain Twp T8 R11 WELS T8 R9 WELS T8 R8 WELS Bloody Brook Saint Croix Stream North Branch Meduxnekeag River W 9 Turner Brook Allagash Stream Millinocket Stream T8 R7 WELS T8 R6 WELS T8 R5 WELS Saint Croix Twp T8 R3 WELS 1 Monticello R Desolation Brook 8 St Francis Brook TC R2 WELS MONTICELLO HOUSING CORP. -

Oyster River Target Fish Community Report.Pdf
NEW HAMPSHIRE STATEWIDE TARGET FISH COMMUNITY ASSESSMENT OYSTER RIVER - FINAL REPORT Prepared for: Prepared by: July, 2018 Table of Contents I. Introduction .................................................................................................................................... 1 II. Designated River Delineation ................................................................................................... 1 Delineation Methods ............................................................................................................................ 1 Delineation Results ............................................................................................................................... 7 III. Reference River Data Selection .............................................................................................. 14 Reference River Selection Methods .................................................................................................... 14 Reference River Selection Results ....................................................................................................... 18 IV. TFC Model Development ........................................................................................................... 28 TFC Model Development Methods ...................................................................................................... 28 TFC Model Results ............................................................................................................................... 29 V. -

Important Northeast Fish Provides Bait & Forage Needs
Species Profile: Atlantic Herring Atlantic Herring Clupea harengus Important Northeast Fish Provides Bait & Forage Needs Introduction Atlantic herring (Clupea harengus) is a member of the clupeid family, which are typically small, schooling marine fishes, such as menhaden, shad, and sardines. This species is also known as sea herring because it spends its entire life cycle in the ASMFC Management Area: ME - NJ ocean (unlike the anadromous river herring). Atlantic herring inhabits the coastal Common Names: Sea herring, sar- waters of the United States from Cape Hatteras, North Carolina through Labra- dine, herring dor, Canada, and also off the coasts of Europe. Herring form the base of the food web as a forage fish for marine mammals, seabirds, and many fish throughout the Interesting Facts: * Atlantic sea herring are often Mid-Atlantic and Northeast. They are an effective and affordable bait source for confused with river herring. Sea lobster, blue crab, and tuna fishermen, and were historically sold by fish canneries herring spend their entire life at as sardines. Whale watching/ecotourism and salt retailers are indirectly dependent sea, while river herring migrate on a steady supply of herring because whales migrate inshore in pursuit of schooling annually to freshwater to spawn. * Atlantic and Pacific herring have herring and fishermen buy salt to preserve their fish. Overseas, frozen and salted been found to produce a burst herring are a valued commodity. of sound, called a Fast Repetitive Tick, at night. Its believed that The Commission’s Atlantic Herring Section manages herring in state waters (0 - 3 this high-pitched click-like sound miles from shore), while the New England Fishery Management Council (Council) is used by herring to signal their location, thereby making it easier regulates the stock in federal waters (3 - 200 miles from shore). -

Alewives and Blueback Herring Juila Beaty University of Maine
The University of Maine DigitalCommons@UMaine Maine Sea Grant Publications Maine Sea Grant 2014 Fisheries Then: Alewives and Blueback Herring Juila Beaty University of Maine Follow this and additional works at: https://digitalcommons.library.umaine.edu/seagrant_pub Part of the Aquaculture and Fisheries Commons Repository Citation Beaty, Juila, "Fisheries Then: Alewives and Blueback Herring" (2014). Maine Sea Grant Publications. 71. https://digitalcommons.library.umaine.edu/seagrant_pub/71 This Article is brought to you for free and open access by DigitalCommons@UMaine. It has been accepted for inclusion in Maine Sea Grant Publications by an authorized administrator of DigitalCommons@UMaine. For more information, please contact [email protected]. (http://www.downeastfisheriestrail.org) Alewives and Blueback Herring Fisheries Then: Alewives and Blueback Herring (i.e. River Herring) By Julia Beaty and Natalie Springuel Reviewed by Chris Bartlett, Dan Kircheis The term “river herring” collectively refers to two species: Alosa pseudoharengus, commonly known as alewife, and the closely related Alosa aestivalis, commonly known as blueback herring, or simply bluebacks. Records dating back to the early nineteenth century indicate that fishermen could tell the difference between alewives and bluebacks, which look very similar; however, historically they have been harvested together with little regard to the differences between the two (Collette and KleinMacPhee 2002). Alewives are the more common of the two species in most rivers in Maine (Collette and Klien MacPhee 2002). Fishermen in Maine often use the word “alewife” to refer to both alewives and bluebacks. Both alewives and bluebacks are anadromous fish, meaning that they are born in fresh water, but spend the majority of their adult lives at sea. -

Atlantic Herring
Species Profile: Atlantic Herring New Stock Assessment Could Lead to Species Snapshot Management Changes Introduction Atlantic Herring Until recently, the Atlantic herring stock had been considered healthy and fully rebuilt from a Clupea harengus collapsed stock in the 1980s. However, the results of the 2018 benchmark stock assessment have raised new concerns about the Atlantic herring resource. While the stock remains not Management Unit: Maine through New Jersey overfished and was not experiencing overfishing in the terminal year (2017) of the assess- ment, the assessment did show very low levels of recruitment over the past five years. These Common Names: Sea herring, sardine, sild, results will likely have management implications for the species as regulators work to prevent common herring, Labrador herring, sperling overfishing from occurring in the coming years. Diminished stock size and, in turn, lowered catch limits will also impact fisheries that rely on Atlantic herring as an important source of Interesting Facts: bait, such as American lobster, blue crab, tuna, and striped bass fisheries. • Atlantic herring and other clupeid fish have exceptional hearing. They can detect sound Life History frequencies up to 40 kilohertz, beyond the Atlantic sea herring is one of 200 species in the clupeid family, which includes menhaden, range of most fish. This allows schooling fish shad, and river herring. It inhabits coastal waters of the U.S. from Cape Hatteras, North Caro- to communicate while avoiding detection by lina through Labrador, Canada, and off the coast of Europe. Herring form the base of the food predatory fish. web as a forage species for many animals, from starfish and whelk to economically import- • While most members of the clupeid family are ant fish such as haddock, cod, and flounder. -

Fish Passage Engineering Design Criteria 2019
FISH PASSAGE ENGINEERING DESIGN CRITERIA 2019 37.2’ U.S. Fish and Wildlife Service Northeast Region June 2019 Fish and Aquatic Conservation, Fish Passage Engineering Ecological Services, Conservation Planning Assistance United States Fish and Wildlife Service Region 5 FISH PASSAGE ENGINEERING DESIGN CRITERIA June 2019 This manual replaces all previous editions of the Fish Passage Engineering Design Criteria issued by the U.S. Fish and Wildlife Service Region 5 Suggested citation: USFWS (U.S. Fish and Wildlife Service). 2019. Fish Passage Engineering Design Criteria. USFWS, Northeast Region R5, Hadley, Massachusetts. USFWS R5 Fish Passage Engineering Design Criteria June 2019 USFWS R5 Fish Passage Engineering Design Criteria June 2019 Contents List of Figures ................................................................................................................................ ix List of Tables .................................................................................................................................. x List of Equations ............................................................................................................................ xi List of Appendices ........................................................................................................................ xii 1 Scope of this Document ....................................................................................................... 1-1 1.1 Role of the USFWS Region 5 Fish Passage Engineering ............................................ -

An Assessment of Nitrogen and Phosphorus Availability in Constructed Wetlands in the Cumberland Marsh Region, Canada
An assessment of nitrogen and phosphorus availability in constructed wetlands in the Cumberland Marsh Region, Canada by Maxwell J. Turner Thesis in partial fulfilment of the requirements for the Degree of Bachelor of Science with Honours in Geology Acadia University April, 2016 © Copyright by Maxwell J. Turner 2016 The thesis by Maxwell J. Turner is accepted in its present form by the Department of Earth and Environmental Science as satisfying the thesis requirements for the degree of Bachelor of Science with Honours Approved by Thesis Supervisors _____________________________ ______________________________ Dr. Ian Spooner Date _____________________________ ______________________________ Dr. Mark Mallory Date Approved by the Head of the Department _____________________________ ______________________________ Dr. Ian Spooner Date Approved by the Honours Committee ______________________________ _______________________________ Dr. Anna Redden Date ii I, Max Turner, grant permission to the University Librarian at Acadia University to reproduce, loan or distribute copies of my thesis in microform, paper or electronic formats on a non-profit basis. I, however, retain copyright in my thesis. ___________________________________ Maxwell Turner ___________________________________ Date iii Acknowledgements I would like to extend recognition to Acadia University and Ducks Unlimited Canada, whose funding and dedication to scientific research made this project possible. Nic McLellan of Ducks Unlimited provided both in-field help and a useful supply of regional knowledge. I would like to thank the entirety of the Department of Earth and Environmental Science for providing a supportive learning environment that allows one to feel comfortable, acknowledged, and feel the expectation for success; but a special thanks to Dr. Rob Raeside whose subtle acknowledgements truly made me feel that this was the department to which I belonged. -

A History of Oysters in Maine (1600S-1970S) Randy Lackovic University of Maine, [email protected]
The University of Maine DigitalCommons@UMaine Darling Marine Center Historical Documents Darling Marine Center Historical Collections 3-2019 A History of Oysters in Maine (1600s-1970s) Randy Lackovic University of Maine, [email protected] Follow this and additional works at: https://digitalcommons.library.umaine.edu/dmc_documents Part of the Aquaculture and Fisheries Commons, History of Science, Technology, and Medicine Commons, and the United States History Commons Repository Citation Lackovic, Randy, "A History of Oysters in Maine (1600s-1970s)" (2019). Darling Marine Center Historical Documents. 22. https://digitalcommons.library.umaine.edu/dmc_documents/22 This Newsletter is brought to you for free and open access by DigitalCommons@UMaine. It has been accepted for inclusion in Darling Marine Center Historical Documents by an authorized administrator of DigitalCommons@UMaine. For more information, please contact [email protected]. A History of Oysters in Maine (1600s-1970s) This is a history of oyster abundance in Maine, and the subsequent decline of oyster abundance. It is a history of oystering, oyster fisheries, and oyster commerce in Maine. It is a history of the transplanting of oysters to Maine, and experiments with oysters in Maine, and of oyster culture in Maine. This history takes place from the 1600s to the 1970s. 17th Century {}{}{}{} In early days, oysters were to be found in lavish abundance along all the Atlantic coast, though Ingersoll says it was at least a small number of oysters on the Gulf of Maine coast.86, 87 Champlain wrote that in 1604, "All the harbors, bays, and coasts from Chouacoet (Saco) are filled with every variety of fish. -
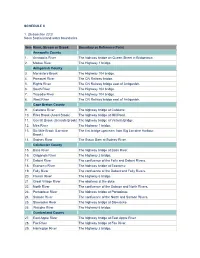
Nova Scotia Inland Water Boundaries Item River, Stream Or Brook
SCHEDULE II 1. (Subsection 2(1)) Nova Scotia inland water boundaries Item River, Stream or Brook Boundary or Reference Point Annapolis County 1. Annapolis River The highway bridge on Queen Street in Bridgetown. 2. Moose River The Highway 1 bridge. Antigonish County 3. Monastery Brook The Highway 104 bridge. 4. Pomquet River The CN Railway bridge. 5. Rights River The CN Railway bridge east of Antigonish. 6. South River The Highway 104 bridge. 7. Tracadie River The Highway 104 bridge. 8. West River The CN Railway bridge east of Antigonish. Cape Breton County 9. Catalone River The highway bridge at Catalone. 10. Fifes Brook (Aconi Brook) The highway bridge at Mill Pond. 11. Gerratt Brook (Gerards Brook) The highway bridge at Victoria Bridge. 12. Mira River The Highway 1 bridge. 13. Six Mile Brook (Lorraine The first bridge upstream from Big Lorraine Harbour. Brook) 14. Sydney River The Sysco Dam at Sydney River. Colchester County 15. Bass River The highway bridge at Bass River. 16. Chiganois River The Highway 2 bridge. 17. Debert River The confluence of the Folly and Debert Rivers. 18. Economy River The highway bridge at Economy. 19. Folly River The confluence of the Debert and Folly Rivers. 20. French River The Highway 6 bridge. 21. Great Village River The aboiteau at the dyke. 22. North River The confluence of the Salmon and North Rivers. 23. Portapique River The highway bridge at Portapique. 24. Salmon River The confluence of the North and Salmon Rivers. 25. Stewiacke River The highway bridge at Stewiacke. 26. Waughs River The Highway 6 bridge. -

Little Fish, Big Impact: Managing a Crucial Link in Ocean Food Webs
little fish BIG IMPACT Managing a crucial link in ocean food webs A report from the Lenfest Forage Fish Task Force The Lenfest Ocean Program invests in scientific research on the environmental, economic, and social impacts of fishing, fisheries management, and aquaculture. Supported research projects result in peer-reviewed publications in leading scientific journals. The Program works with the scientists to ensure that research results are delivered effectively to decision makers and the public, who can take action based on the findings. The program was established in 2004 by the Lenfest Foundation and is managed by the Pew Charitable Trusts (www.lenfestocean.org, Twitter handle: @LenfestOcean). The Institute for Ocean Conservation Science (IOCS) is part of the Stony Brook University School of Marine and Atmospheric Sciences. It is dedicated to advancing ocean conservation through science. IOCS conducts world-class scientific research that increases knowledge about critical threats to oceans and their inhabitants, provides the foundation for smarter ocean policy, and establishes new frameworks for improved ocean conservation. Suggested citation: Pikitch, E., Boersma, P.D., Boyd, I.L., Conover, D.O., Cury, P., Essington, T., Heppell, S.S., Houde, E.D., Mangel, M., Pauly, D., Plagányi, É., Sainsbury, K., and Steneck, R.S. 2012. Little Fish, Big Impact: Managing a Crucial Link in Ocean Food Webs. Lenfest Ocean Program. Washington, DC. 108 pp. Cover photo illustration: shoal of forage fish (center), surrounded by (clockwise from top), humpback whale, Cape gannet, Steller sea lions, Atlantic puffins, sardines and black-legged kittiwake. Credits Cover (center) and title page: © Jason Pickering/SeaPics.com Banner, pages ii–1: © Brandon Cole Design: Janin/Cliff Design Inc. -

South Western Nova Scotia
Netukulimk of Aquatic Natural Life “The N.C.N.S. Netukulimkewe’l Commission is the Natural Life Management Authority for the Large Community of Mi’kmaq /Aboriginal Peoples who continue to reside on Traditional Mi’Kmaq Territory in Nova Scotia undisplaced to Indian Act Reserves” P.O. Box 1320, Truro, N.S., B2N 5N2 Tel: 902-895-7050 Toll Free: 1-877-565-1752 2 Netukulimk of Aquatic Natural Life N.C.N.S. Netukulimkewe’l Commission Table of Contents: Page(s) The 1986 Proclamation by our late Mi’kmaq Grand Chief 4 The 1994 Commendation to all A.T.R.A. Netukli’tite’wk (Harvesters) 5 A Message From the N.C.N.S. Netukulimkewe’l Commission 6 Our Collective Rights Proclamation 7 A.T.R.A. Netukli’tite’wk (Harvester) Duties and Responsibilities 8-12 SCHEDULE I Responsible Netukulimkewe’l (Harvesting) Methods and Equipment 16 Dangers of Illegal Harvesting- Enjoy Safe Shellfish 17-19 Anglers Guide to Fishes Of Nova Scotia 20-21 SCHEDULE II Specific Species Exceptions 22 Mntmu’k, Saqskale’s, E’s and Nkata’laq (Oysters, Scallops, Clams and Mussels) 22 Maqtewe’kji’ka’w (Small Mouth Black Bass) 23 Elapaqnte’mat Ji’ka’w (Striped Bass) 24 Atoqwa’su (Trout), all types 25 Landlocked Plamu (Landlocked Salmon) 26 WenjiWape’k Mime’j (Atlantic Whitefish) 26 Lake Whitefish 26 Jakej (Lobster) 27 Other Species 33 Atlantic Plamu (Salmon) 34 Atlantic Plamu (Salmon) Netukulimk (Harvest) Zones, Seasons and Recommended Netukulimk (Harvest) Amounts: 55 SCHEDULE III Winter Lake Netukulimkewe’l (Harvesting) 56-62 Fishing and Water Safety 63 Protecting Our Community’s Aboriginal and Treaty Rights-Community 66-70 Dispositions and Appeals Regional Netukulimkewe’l Advisory Councils (R.N.A.C.’s) 74-75 Description of the 2018 N.C.N.S. -

Our Maritime Heritage a Piscataqua Region Timeline
OUR MARITIME HERITAGE A PISCATAQUA REGION TIMELINE 14,000 years ago Glaciers melted 8,000 years ago Evidence of seasonal human activity along the Lamprey River 2,000 years ago Sea level reached today’s current levels 9approximately) Before 1600 Native Americans had been in area for thousands of years Early 1400s Evidence of farming by Natives in Eliot 1500s European explorers and fishermen visiting and trading in region 1524 Verrazano became first European to describe the Maine coast Early 1600s English settlements at Exeter, Dover, Hampton, and Kittery Early 1600s Native population devastated by European diseases 1602 Earliest landfall on the coast in York (claimed) 1607 Popham Colony established at Maine’s Kennebec River; lasts barely a year 1603 Martin Pring arrived, looking for sassafras FISHING, BEAVER TRADE 1614 Captain John Smith created the first map of the region 1620 Pilgrims from the MAYFLOWER settled at Plimoth in Massachusetts Bay 1622-23 King James granted charters to Mason and Georges for Piscataqua Plantations 1623 Fishing settlements established at Odiorne Point and Dover (Hilton) Point 1623 Kittery area is settled; incorporated in 1647, billed as oldest town in Maine 1623 Simple earthen defense was built at Fort Point (later Fort William and Mary) 1624 Captain Christopher Levitt sailed up the York River 1630 Strawbery Banke settled by Captain Neal and band of Englishmen 1630 Europeans first settle below the falls on the Salmon Falls River 1631 Stratham settled by Europeans under Captain Thomas Wiggin 1632 Fort William