Copy Number Variants in Attention-Deficit Hyperactive Disorder
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Protein Identities in Evs Isolated from U87-MG GBM Cells As Determined by NG LC-MS/MS
Protein identities in EVs isolated from U87-MG GBM cells as determined by NG LC-MS/MS. No. Accession Description Σ Coverage Σ# Proteins Σ# Unique Peptides Σ# Peptides Σ# PSMs # AAs MW [kDa] calc. pI 1 A8MS94 Putative golgin subfamily A member 2-like protein 5 OS=Homo sapiens PE=5 SV=2 - [GG2L5_HUMAN] 100 1 1 7 88 110 12,03704523 5,681152344 2 P60660 Myosin light polypeptide 6 OS=Homo sapiens GN=MYL6 PE=1 SV=2 - [MYL6_HUMAN] 100 3 5 17 173 151 16,91913397 4,652832031 3 Q6ZYL4 General transcription factor IIH subunit 5 OS=Homo sapiens GN=GTF2H5 PE=1 SV=1 - [TF2H5_HUMAN] 98,59 1 1 4 13 71 8,048185945 4,652832031 4 P60709 Actin, cytoplasmic 1 OS=Homo sapiens GN=ACTB PE=1 SV=1 - [ACTB_HUMAN] 97,6 5 5 35 917 375 41,70973209 5,478027344 5 P13489 Ribonuclease inhibitor OS=Homo sapiens GN=RNH1 PE=1 SV=2 - [RINI_HUMAN] 96,75 1 12 37 173 461 49,94108966 4,817871094 6 P09382 Galectin-1 OS=Homo sapiens GN=LGALS1 PE=1 SV=2 - [LEG1_HUMAN] 96,3 1 7 14 283 135 14,70620005 5,503417969 7 P60174 Triosephosphate isomerase OS=Homo sapiens GN=TPI1 PE=1 SV=3 - [TPIS_HUMAN] 95,1 3 16 25 375 286 30,77169764 5,922363281 8 P04406 Glyceraldehyde-3-phosphate dehydrogenase OS=Homo sapiens GN=GAPDH PE=1 SV=3 - [G3P_HUMAN] 94,63 2 13 31 509 335 36,03039959 8,455566406 9 Q15185 Prostaglandin E synthase 3 OS=Homo sapiens GN=PTGES3 PE=1 SV=1 - [TEBP_HUMAN] 93,13 1 5 12 74 160 18,68541938 4,538574219 10 P09417 Dihydropteridine reductase OS=Homo sapiens GN=QDPR PE=1 SV=2 - [DHPR_HUMAN] 93,03 1 1 17 69 244 25,77302971 7,371582031 11 P01911 HLA class II histocompatibility antigen, -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Cldn19 Clic2 Clmp Cln3
NewbornDx™ Advanced Sequencing Evaluation When time to diagnosis matters, the NewbornDx™ Advanced Sequencing Evaluation from Athena Diagnostics delivers rapid, 5- to 7-day results on a targeted 1,722-genes. A2ML1 ALAD ATM CAV1 CLDN19 CTNS DOCK7 ETFB FOXC2 GLUL HOXC13 JAK3 AAAS ALAS2 ATP1A2 CBL CLIC2 CTRC DOCK8 ETFDH FOXE1 GLYCTK HOXD13 JUP AARS2 ALDH18A1 ATP1A3 CBS CLMP CTSA DOK7 ETHE1 FOXE3 GM2A HPD KANK1 AASS ALDH1A2 ATP2B3 CC2D2A CLN3 CTSD DOLK EVC FOXF1 GMPPA HPGD K ANSL1 ABAT ALDH3A2 ATP5A1 CCDC103 CLN5 CTSK DPAGT1 EVC2 FOXG1 GMPPB HPRT1 KAT6B ABCA12 ALDH4A1 ATP5E CCDC114 CLN6 CUBN DPM1 EXOC4 FOXH1 GNA11 HPSE2 KCNA2 ABCA3 ALDH5A1 ATP6AP2 CCDC151 CLN8 CUL4B DPM2 EXOSC3 FOXI1 GNAI3 HRAS KCNB1 ABCA4 ALDH7A1 ATP6V0A2 CCDC22 CLP1 CUL7 DPM3 EXPH5 FOXL2 GNAO1 HSD17B10 KCND2 ABCB11 ALDOA ATP6V1B1 CCDC39 CLPB CXCR4 DPP6 EYA1 FOXP1 GNAS HSD17B4 KCNE1 ABCB4 ALDOB ATP7A CCDC40 CLPP CYB5R3 DPYD EZH2 FOXP2 GNE HSD3B2 KCNE2 ABCB6 ALG1 ATP8A2 CCDC65 CNNM2 CYC1 DPYS F10 FOXP3 GNMT HSD3B7 KCNH2 ABCB7 ALG11 ATP8B1 CCDC78 CNTN1 CYP11B1 DRC1 F11 FOXRED1 GNPAT HSPD1 KCNH5 ABCC2 ALG12 ATPAF2 CCDC8 CNTNAP1 CYP11B2 DSC2 F13A1 FRAS1 GNPTAB HSPG2 KCNJ10 ABCC8 ALG13 ATR CCDC88C CNTNAP2 CYP17A1 DSG1 F13B FREM1 GNPTG HUWE1 KCNJ11 ABCC9 ALG14 ATRX CCND2 COA5 CYP1B1 DSP F2 FREM2 GNS HYDIN KCNJ13 ABCD3 ALG2 AUH CCNO COG1 CYP24A1 DST F5 FRMD7 GORAB HYLS1 KCNJ2 ABCD4 ALG3 B3GALNT2 CCS COG4 CYP26C1 DSTYK F7 FTCD GP1BA IBA57 KCNJ5 ABHD5 ALG6 B3GAT3 CCT5 COG5 CYP27A1 DTNA F8 FTO GP1BB ICK KCNJ8 ACAD8 ALG8 B3GLCT CD151 COG6 CYP27B1 DUOX2 F9 FUCA1 GP6 ICOS KCNK3 ACAD9 ALG9 -

Effects on Steroid 5-Alpha Reductase Gene Expression of Thai Rice Bran Extracts and Molecular Dynamics Study on SRD5A2
biology Article Effects on Steroid 5-Alpha Reductase Gene Expression of Thai Rice Bran Extracts and Molecular Dynamics Study on SRD5A2 Chiranan Khantham 1 , Wipawadee Yooin 1,2 , Korawan Sringarm 2,3 , Sarana Rose Sommano 2,4 , Supat Jiranusornkul 1, Francisco David Carmona 5,6 , Wutigri Nimlamool 7 , Pensak Jantrawut 1,2, Pornchai Rachtanapun 8,9 and Warintorn Ruksiriwanich 1,2,8,* 1 Department of Pharmaceutical Sciences, Faculty of Pharmacy, Chiang Mai University, Chiang Mai 50200, Thailand; [email protected] (C.K.); [email protected] (W.Y.); [email protected] (S.J.); [email protected] (P.J.) 2 Cluster of Research and Development of Pharmaceutical and Natural Products Innovation for Human or Animal, Chiang Mai University, Chiang Mai 50200, Thailand; [email protected] (K.S.); [email protected] (S.R.S.) 3 Department of Animal and Aquatic Sciences, Faculty of Agriculture, Chiang Mai University, Chiang Mai 50200, Thailand 4 Plant Bioactive Compound Laboratory (BAC), Department of Plant and Soil Sciences, Faculty of Agriculture, Chiang Mai University, Chiang Mai 50200, Thailand 5 Departamento de Genética e Instituto de Biotecnología, Universidad de Granada, 18071 Granada, Spain; [email protected] 6 Instituto de Investigación Biosanitaria ibs.GRANADA, 18014 Granada, Spain 7 Citation: Khantham, C.; Yooin, W.; Department of Pharmacology, Faculty of Medicine, Chiang Mai University, Chiang Mai 50200, Thailand; Sringarm, K.; Sommano, S.R.; [email protected] 8 Cluster of Agro Bio-Circular-Green Industry, Faculty of Agro-Industry, Chiang Mai University, Jiranusornkul, S.; Carmona, F.D.; Chiang Mai 50100, Thailand; [email protected] Nimlamool, W.; Jantrawut, P.; 9 School of Agro-Industry, Faculty of Agro-Industry, Chiang Mai University, Chiang Mai 50100, Thailand Rachtanapun, P.; Ruksiriwanich, W. -

Molecular Diagnostic Requisition
BAYLOR MIRACA GENETICS LABORATORIES SHIP TO: Baylor Miraca Genetics Laboratories 2450 Holcombe, Grand Blvd. -Receiving Dock PHONE: 800-411-GENE | FAX: 713-798-2787 | www.bmgl.com Houston, TX 77021-2024 Phone: 713-798-6555 MOLECULAR DIAGNOSTIC REQUISITION PATIENT INFORMATION SAMPLE INFORMATION NAME: DATE OF COLLECTION: / / LAST NAME FIRST NAME MI MM DD YY HOSPITAL#: ACCESSION#: DATE OF BIRTH: / / GENDER (Please select one): FEMALE MALE MM DD YY SAMPLE TYPE (Please select one): ETHNIC BACKGROUND (Select all that apply): UNKNOWN BLOOD AFRICAN AMERICAN CORD BLOOD ASIAN SKELETAL MUSCLE ASHKENAZIC JEWISH MUSCLE EUROPEAN CAUCASIAN -OR- DNA (Specify Source): HISPANIC NATIVE AMERICAN INDIAN PLACE PATIENT STICKER HERE OTHER JEWISH OTHER (Specify): OTHER (Please specify): REPORTING INFORMATION ADDITIONAL PROFESSIONAL REPORT RECIPIENTS PHYSICIAN: NAME: INSTITUTION: PHONE: FAX: PHONE: FAX: NAME: EMAIL (INTERNATIONAL CLIENT REQUIREMENT): PHONE: FAX: INDICATION FOR STUDY SYMPTOMATIC (Summarize below.): *FAMILIAL MUTATION/VARIANT ANALYSIS: COMPLETE ALL FIELDS BELOW AND ATTACH THE PROBAND'S REPORT. GENE NAME: ASYMPTOMATIC/POSITIVE FAMILY HISTORY: (ATTACH FAMILY HISTORY) MUTATION/UNCLASSIFIED VARIANT: RELATIONSHIP TO PROBAND: THIS INDIVIDUAL IS CURRENTLY: SYMPTOMATIC ASYMPTOMATIC *If family mutation is known, complete the FAMILIAL MUTATION/ VARIANT ANALYSIS section. NAME OF PROBAND: ASYMPTOMATIC/POPULATION SCREENING RELATIONSHIP TO PROBAND: OTHER (Specify clinical findings below): BMGL LAB#: A COPY OF ORIGINAL RESULTS ATTACHED IF PROBAND TESTING WAS PERFORMED AT ANOTHER LAB, CALL TO DISCUSS PRIOR TO SENDING SAMPLE. A POSITIVE CONTROL MAY BE REQUIRED IN SOME CASES. REQUIRED: NEW YORK STATE PHYSICIAN SIGNATURE OF CONSENT I certify that the patient specified above and/or their legal guardian has been informed of the benefits, risks, and limitations of the laboratory test(s) requested. -

Genetic Background of Ataxia in Children Younger Than 5 Years in Finland E444
Volume 6, Number 4, August 2020 Neurology.org/NG A peer-reviewed clinical and translational neurology open access journal ARTICLE Genetic background of ataxia in children younger than 5 years in Finland e444 ARTICLE Cerebral arteriopathy associated with heterozygous variants in the casitas B-lineage lymphoma gene e448 ARTICLE Somatic SLC35A2 mosaicism correlates with clinical fi ndings in epilepsy brain tissuee460 ARTICLE Synonymous variants associated with Alzheimer disease in multiplex families e450 Academy Officers Neurology® is a registered trademark of the American Academy of Neurology (registration valid in the United States). James C. Stevens, MD, FAAN, President Neurology® Genetics (eISSN 2376-7839) is an open access journal published Orly Avitzur, MD, MBA, FAAN, President Elect online for the American Academy of Neurology, 201 Chicago Avenue, Ann H. Tilton, MD, FAAN, Vice President Minneapolis, MN 55415, by Wolters Kluwer Health, Inc. at 14700 Citicorp Drive, Bldg. 3, Hagerstown, MD 21742. Business offices are located at Two Carlayne E. Jackson, MD, FAAN, Secretary Commerce Square, 2001 Market Street, Philadelphia, PA 19103. Production offices are located at 351 West Camden Street, Baltimore, MD 21201-2436. Janis M. Miyasaki, MD, MEd, FRCPC, FAAN, Treasurer © 2020 American Academy of Neurology. Ralph L. Sacco, MD, MS, FAAN, Past President Neurology® Genetics is an official journal of the American Academy of Neurology. Journal website: Neurology.org/ng, AAN website: AAN.com CEO, American Academy of Neurology Copyright and Permission Information: Please go to the journal website (www.neurology.org/ng) and click the Permissions tab for the relevant Mary E. Post, MBA, CAE article. Alternatively, send an email to [email protected]. -

Suppression of Steroid 5Α-Reductase Type I Promotes Cellular Apoptosis
Dou et al. Cell Death and Disease (2021) 12:206 https://doi.org/10.1038/s41419-021-03510-4 Cell Death & Disease ARTICLE Open Access Suppression of steroid 5α-reductase type I promotes cellular apoptosis and autophagy via PI3K/Akt/mTOR pathway in multiple myeloma Renjie Dou1,2, Jinjun Qian2,WeiWu1,YanxinZhang2,YuxiaYuan2, Mengjie Guo1,2,RongfangWei2, Shu Yang2, Artur Jurczyszyn 3, Siegfried Janz 4, Meral Beksac5, Chunyan Gu 1,2 and Ye Yang1,2,6 Abstract Steroid 5α-reductase type I (SRD5A1) is a validated oncogene in many sex hormone-related cancers, but its role in multiple myeloma (MM) remains unknown. Based on gene expression profiling (GEP) of sequential MM samples during the disease course, we found that the aberrant expression of SRD5A1 was correlated with progression and poor prognosis in MM patients. In this study, the oncogenic roles of SRD5A1 were validated in human MM cell lines (ARP1 and H929) and the xenograft MM model as well as the 5TMM mouse model. MTT and flow cytometry were used to assess MM cell proliferation, cell cycle, and apoptosis post inducible knockdown SRD5A1 by lentivirus-mediated short- hairpin RNA (shRNA). Transcriptomic sequencing, immunofluorescence, and western blot were used to investigate the effects of SRD5A1 suppression on cell apoptosis and autophagy. Mechanistically, SRD5A1 downregulation simultaneously regulated both the Bcl-2 family protein-mediated apoptosis and the autophagic process via PI3K/Akt/ mTOR signaling pathway in MM cells. Meanwhile, the autophagy inhibitor (3-methyladenine) and SRD5A1 inhibitor (Dutasteride) were utilized to evaluate their anti-myeloma effect. Thus, our results demonstrated that SRD5A1 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; downregulation simultaneously regulated both the apoptosis and the autophagic process in MM cells. -
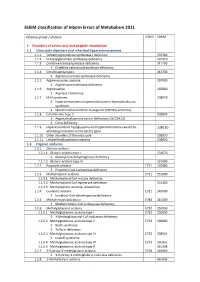
SSIEM Classification of Inborn Errors of Metabolism 2011
SSIEM classification of Inborn Errors of Metabolism 2011 Disease group / disease ICD10 OMIM 1. Disorders of amino acid and peptide metabolism 1.1. Urea cycle disorders and inherited hyperammonaemias 1.1.1. Carbamoylphosphate synthetase I deficiency 237300 1.1.2. N-Acetylglutamate synthetase deficiency 237310 1.1.3. Ornithine transcarbamylase deficiency 311250 S Ornithine carbamoyltransferase deficiency 1.1.4. Citrullinaemia type1 215700 S Argininosuccinate synthetase deficiency 1.1.5. Argininosuccinic aciduria 207900 S Argininosuccinate lyase deficiency 1.1.6. Argininaemia 207800 S Arginase I deficiency 1.1.7. HHH syndrome 238970 S Hyperammonaemia-hyperornithinaemia-homocitrullinuria syndrome S Mitochondrial ornithine transporter (ORNT1) deficiency 1.1.8. Citrullinemia Type 2 603859 S Aspartate glutamate carrier deficiency ( SLC25A13) S Citrin deficiency 1.1.9. Hyperinsulinemic hypoglycemia and hyperammonemia caused by 138130 activating mutations in the GLUD1 gene 1.1.10. Other disorders of the urea cycle 238970 1.1.11. Unspecified hyperammonaemia 238970 1.2. Organic acidurias 1.2.1. Glutaric aciduria 1.2.1.1. Glutaric aciduria type I 231670 S Glutaryl-CoA dehydrogenase deficiency 1.2.1.2. Glutaric aciduria type III 231690 1.2.2. Propionic aciduria E711 232000 S Propionyl-CoA-Carboxylase deficiency 1.2.3. Methylmalonic aciduria E711 251000 1.2.3.1. Methylmalonyl-CoA mutase deficiency 1.2.3.2. Methylmalonyl-CoA epimerase deficiency 251120 1.2.3.3. Methylmalonic aciduria, unspecified 1.2.4. Isovaleric aciduria E711 243500 S Isovaleryl-CoA dehydrogenase deficiency 1.2.5. Methylcrotonylglycinuria E744 210200 S Methylcrotonyl-CoA carboxylase deficiency 1.2.6. Methylglutaconic aciduria E712 250950 1.2.6.1. Methylglutaconic aciduria type I E712 250950 S 3-Methylglutaconyl-CoA hydratase deficiency 1.2.6.2. -

Spatial Sorting Enables Comprehensive Characterization of Liver Zonation
ARTICLES https://doi.org/10.1038/s42255-019-0109-9 Spatial sorting enables comprehensive characterization of liver zonation Shani Ben-Moshe1,3, Yonatan Shapira1,3, Andreas E. Moor 1,2, Rita Manco1, Tamar Veg1, Keren Bahar Halpern1 and Shalev Itzkovitz 1* The mammalian liver is composed of repeating hexagonal units termed lobules. Spatially resolved single-cell transcriptomics has revealed that about half of hepatocyte genes are differentially expressed across the lobule, yet technical limitations have impeded reconstructing similar global spatial maps of other hepatocyte features. Here, we show how zonated surface markers can be used to sort hepatocytes from defined lobule zones with high spatial resolution. We apply transcriptomics, microRNA (miRNA) array measurements and mass spectrometry proteomics to reconstruct spatial atlases of multiple zon- ated features. We demonstrate that protein zonation largely overlaps with messenger RNA zonation, with the periportal HNF4α as an exception. We identify zonation of miRNAs, such as miR-122, and inverse zonation of miRNAs and their hepa- tocyte target genes, highlighting potential regulation of gene expression levels through zonated mRNA degradation. Among the targets, we find the pericentral Wingless-related integration site (Wnt) receptors Fzd7 and Fzd8 and the periportal Wnt inhibitors Tcf7l1 and Ctnnbip1. Our approach facilitates reconstructing spatial atlases of multiple cellular features in the liver and other structured tissues. he mammalian liver is a structured organ, consisting of measurements would broaden our understanding of the regulation repeating hexagonally shaped units termed ‘lobules’ (Fig. 1a). of liver zonation and could be used to model liver metabolic func- In mice, each lobule consists of around 9–12 concentric lay- tion more precisely. -

Structure of Human Steroid 5Α-Reductase 2 With
Structure of human steroid 5α-reductase 2 with anti-androgen drug finasteride Qingpin Xiao1, 5, Lei Wang2, Shreyas Supekar3, Tao Shen4, Heng Liu2, Fei Ye4, Junzhou Huang4, Hao Fan3, Zhiyi Wei1*, Cheng Zhang2* 1Department of Biology, Southern University of Science and Technology, Shenzhen, Guangdong 518055, China 2Department of Pharmacology and Chemical Biology, School of Medicine, University of Pittsburgh, Pittsburgh, PA15261, USA 3Bioinformatics Institute (BII), Agency for Science, Technology and Research (A*STAR), Singapore 138671, Singapore 4Tencent AI Lab, Shenzhen, Guangdong 518000, China 5Faculty of Health Sciences, University of Macau, Macau SAR 999078, China. *Correspondence: Dr. Zhiyi Wei, [email protected] or Dr. Cheng Zhang, [email protected]. 1 Abstract Human steroid 5α-reductase 2 (SRD5α2) as a critical integral membrane enzyme in steroid metabolism catalyzes testosterone to dihydrotestosterone. Mutations on its gene have been linked to 5α-reductase deficiency and prostate cancer. Finasteride and dutasteride as SRD5α2 inhibitors are widely used anti- androgen drugs for benign prostate hyperplasia, which have recently been indicated in the treatment of COVID-19. The molecular mechanisms underlying enzyme catalysis and inhibition remained elusive for SRD5α2 and other eukaryotic integral membrane steroid reductases due to a lack of structural information. Here, we report a crystal structure of human SRD5α2 at 2.8 Å revealing a unique 7-TM structural topology and an intermediate adduct of finasteride and NADPH as NADP- dihydrofinasteride in a largely enclosed binding cavity inside the membrane. Structural analysis together with computational and mutagenesis studies reveals molecular mechanisms for the 5α- reduction of testosterone and the finasteride inhibition involving residues E57 and Y91. -

Quantitative Trait Loci Mapping of Macrophage Atherogenic Phenotypes
QUANTITATIVE TRAIT LOCI MAPPING OF MACROPHAGE ATHEROGENIC PHENOTYPES BRIAN RITCHEY Bachelor of Science Biochemistry John Carroll University May 2009 submitted in partial fulfillment of requirements for the degree DOCTOR OF PHILOSOPHY IN CLINICAL AND BIOANALYTICAL CHEMISTRY at the CLEVELAND STATE UNIVERSITY December 2017 We hereby approve this thesis/dissertation for Brian Ritchey Candidate for the Doctor of Philosophy in Clinical-Bioanalytical Chemistry degree for the Department of Chemistry and the CLEVELAND STATE UNIVERSITY College of Graduate Studies by ______________________________ Date: _________ Dissertation Chairperson, Johnathan D. Smith, PhD Department of Cellular and Molecular Medicine, Cleveland Clinic ______________________________ Date: _________ Dissertation Committee member, David J. Anderson, PhD Department of Chemistry, Cleveland State University ______________________________ Date: _________ Dissertation Committee member, Baochuan Guo, PhD Department of Chemistry, Cleveland State University ______________________________ Date: _________ Dissertation Committee member, Stanley L. Hazen, MD PhD Department of Cellular and Molecular Medicine, Cleveland Clinic ______________________________ Date: _________ Dissertation Committee member, Renliang Zhang, MD PhD Department of Cellular and Molecular Medicine, Cleveland Clinic ______________________________ Date: _________ Dissertation Committee member, Aimin Zhou, PhD Department of Chemistry, Cleveland State University Date of Defense: October 23, 2017 DEDICATION I dedicate this work to my entire family. In particular, my brother Greg Ritchey, and most especially my father Dr. Michael Ritchey, without whose support none of this work would be possible. I am forever grateful to you for your devotion to me and our family. You are an eternal inspiration that will fuel me for the remainder of my life. I am extraordinarily lucky to have grown up in the family I did, which I will never forget. -

Early Upregulation of AR and Steroidogenesis Enzyme Expression After 3 Months of Androgen-Deprivation Therapy Agus Rizal A
Hamid et al. BMC Urology (2020) 20:71 https://doi.org/10.1186/s12894-020-00627-0 RESEARCH ARTICLE Open Access Early upregulation of AR and steroidogenesis enzyme expression after 3 months of androgen-deprivation therapy Agus Rizal A. H. Hamid1* , Harun W. Kusuma Putra1, Ningrum Paramita Sari2, Putri Diana1, Saras Serani Sesari1, Eka Novita3, Fajar Lamhot Gultom3, Meilania Saraswati3, Budiana Tanurahardja3, Asmarinah2, Rainy Umbas1 and Chaidir A. Mochtar1 Abstract Background: Androgen deprivation therapy (ADT) is a standard treatment for advanced prostate cancer (PCa). However, PCa recurrence and progression rates during ADT are high. Until now, there has been no evidence regarding when progression begins. This study evaluated the gene expression of intraprostatic androgen receptor (AR) and steroidogenic enzymes in the early stages of ADT. Methods: Prostate tissue samples were taken from PCa patients with urinary retention who received ADT (ADT-PCa; n = 10) and were further subgrouped into ADT ≤12 months (n = 4) and ADT > 12 months (n = 6). The ADT-PCa tissues were then compared with BPH (n = 12) and primary (no treatment) PCa tissues (n = 16). mRNA for gene expression analysis of AR and steroidogenic enzymes was extracted from formalin-fixed paraffin embedded (FFPE) tissues and analyzed by real-time PCR. Protein expression was evaluated by immunohistochemistry with specific antibodies. Results: AR gene expression was higher in the ADT-PCa group than in the BPH or primary PCa group. Both the ADT ≤12 and > 12 months subgroups had significantly higher relative gene expression levels of AR (p < 0.01 and 0.03, respectively) than the primary PCa group.