A Large-Scale Clinical Validation Study Using Ncapp Cloud Plus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

UNEP-Tongji University Institute of Environment for Sustainable Development
Master and Doctoral Degree Programmes UNEP-Tongji University Institute of Environment for Sustainable Development ABOUT THE PROGRAMME CAREER PROSPECT • Duration: 2 Years/4 Years The programmes are designed to foster high-end business officials, man- • Sept.2021 - July 2023/ agerial personnel and scientific researchers, offering two-year master Sept. 2021 - July 2025 and four-year doctoral programs for the purpose of educating high-end • Language: English and inter-disciplinary talent working in the applied fields of government, • Tuition fee: 39 000 RMB trade, foreign affairs, agriculture, technology, education, culture and per year health, building intellectual capacity and facilitating the economic and social development from the world. • Limited scholarships are available for further con- Our alumni are currently active in international organizations (such as sultation. the World Bank, UNDP, ESCAP, etc.), national government agencies (such as the Chinese Ministry of Ecology Environment, the Turkish Envi- • The programme consists of ronment and Urban Planning Ministry, the Ministry of Labor and Social multi-disciplines courses, Security of Zambia, Egypt Ministry of Agriculture, Ministry of Environ- academic and cultural field ment, Serbian Ministry of Agriculture and Environment, Ministry of Nat- trips and Chinese lan- ural Resources of Ukraine, Ministry of Environment of Romania, Minis- guage. try of Economy and Trade of Indonesia, Ministry of Agriculture of Zim- babwe, Ministry of Energy of Liberia, Ministry of Environment of Ethio- -

Shuang Zhang
June 2020 Shuang Zhang Department of Economics, University of Colorado Boulder Email: [email protected] Website: https://spot.colorado.edu/~shzh6533/index.html APPOINTMENTS 2020- Associate Professor of Economics (with tenure), University of Colorado Boulder 2013-2020 Assistant Professor of Economics, University of Colorado Boulder 2012-2013 SIEPR Postdoctoral Fellow, Stanford University AFFILIATIONS 2020- Faculty Research Fellow, National Bureau of Economic Research EDUCATION Ph.D. in Economics, Cornell University, 2012 M.A. in Economics, Fudan University, China, 2007 B.A. in Economics (with distinction), Shanghai University of Finance and Economics, China, 2004 RESEARCH INTERESTS Environment and Energy, Health, Development, China PUBLICATIONS “Willingness to Pay for Clean Air: Evidence from Air Purifier Markets in China” (with Koichiro Ito). Journal of Political Economy, 2020, 128 (5): 1627-1672. (Lead Article) “Land Reform and Sex Selection in China” (with Douglas Almond and Hongbin Li). Journal of Political Economy, 2019, 127 (2): 560-585. “The Limits of Political Meritocracy: Screening Bureaucrats Under Imperfect Verifiability” (with Juan Carlos Suárez Serrato and Xiao Yu Wang). Journal of Development Economics, 2019, 140: 223-241. “Quantifying Coal Power Plant Responses to Tighter SO2 Emissions Standards in China” (with Va- lerie Karplus and Douglas Almond). Proceedings of the National Academy of Sciences, 2018, 115 (27): 7004-7009. “The Effects of High School Closure on Education and Labor Market Outcomes in Rural China”. Economic Development and Culture Change, 2018, 67 (1): 171-191. 1 WORKING PAPERS Reforming Inefficient Energy Pricing: Evidence from China (with Koichiro Ito), NBER WP 26853, 2020. Ambiguous Pollution Response to COVID-19 in China (with Douglas Almond and Xinming Du), NBER WP 27086, 2020. -

2019 Year Book.Pdf
2019 Contents Preface / P_05> Overview / P_07> SICA Profile / P_15> Cultural Performances and Exhibitions, 2019 / P_19> Foreign Exchange, 2019 / P_45> Academic Conferences, 2019 / P_67> Summary of Cultural Exchanges and Visits, 2019 / P_77> 「Offerings at the First Day of Year」(detail) by YANG Zhengxin Sea Breeze: Exhibition of Shanghai-Style Calligraphy and Painting Preface This year marks the 70th anniversary of the founding of the People’s Republic of China. Over the past 70 years, the Chinese culture has forged ahead regardless of trials and hardships. In the course of its inheritance and development, the Chinese culture has stepped onto the world stage and found her way under spotlight. The SICA, established in the golden age of reform and opening-up, has been adhering to its mission of “strengthening mutual understanding and friendly cooperation between Shanghai and other countries or regions through international cultural exchanges in various areas, so as to promote the economic development, scientific progress and cultural prosperity of the city” for more than 30 years. It has been exploring new modes of international exchange and has been actively engaging in a variety of international culture exchanges on different levels in broad fields. On behalf of the entire staff of the SICA, I hereby would like to extend our sincere gratitude for the concern and support offered by various levels of government departments, Council members of the SICA, partner agencies and cultural institutions, people from all circles of life, and friends from both home and abroad. To sum up our work in the year 2019, we share in this booklet a collection of illustrated reports on the programs in which we have been involved in the past year. -

GLOBAL HISTORY and NEW POLYCENTRIC APPROACHES Europe, Asia and the Americas in a World Network System Palgrave Studies in Comparative Global History
Foreword by Patrick O’Brien Edited by Manuel Perez Garcia · Lucio De Sousa GLOBAL HISTORY AND NEW POLYCENTRIC APPROACHES Europe, Asia and the Americas in a World Network System Palgrave Studies in Comparative Global History Series Editors Manuel Perez Garcia Shanghai Jiao Tong University Shanghai, China Lucio De Sousa Tokyo University of Foreign Studies Tokyo, Japan This series proposes a new geography of Global History research using Asian and Western sources, welcoming quality research and engag- ing outstanding scholarship from China, Europe and the Americas. Promoting academic excellence and critical intellectual analysis, it offers a rich source of global history research in sub-continental areas of Europe, Asia (notably China, Japan and the Philippines) and the Americas and aims to help understand the divergences and convergences between East and West. More information about this series at http://www.springer.com/series/15711 Manuel Perez Garcia · Lucio De Sousa Editors Global History and New Polycentric Approaches Europe, Asia and the Americas in a World Network System Editors Manuel Perez Garcia Lucio De Sousa Shanghai Jiao Tong University Tokyo University of Foreign Studies Shanghai, China Fuchu, Tokyo, Japan Pablo de Olavide University Seville, Spain Palgrave Studies in Comparative Global History ISBN 978-981-10-4052-8 ISBN 978-981-10-4053-5 (eBook) https://doi.org/10.1007/978-981-10-4053-5 Library of Congress Control Number: 2017937489 © The Editor(s) (if applicable) and The Author(s) 2018, corrected publication 2018. This book is an open access publication. Open Access This book is licensed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license and indicate if changes were made. -

1 Zhirong “Jerry” Zhao
Updated 03/16/2020 ZHIRONG “JERRY” ZHAO Gross Family Professor, Public & Nonprofit Management Hubert H. Humphrey School of Public Affairs, University of Minnesota (UMN) 301 19th Avenue South, Minneapolis, MN 55455 [email protected], 612-625-7318 (phone) BIOGRAPHICAL INFORMATION Education 2005 Ph.D. of Public Administration, University of Georgia (UGA), Athens 1997 Master of Urban Planning, Tongji University, Shanghai, China 1993 Bachelor of Urban Planning, Tongji University, Shanghai, China Academic Positions 2019- pres. Chair Leadership & Management Area, Humphrey School of Public Affairs, UMN 2019–pres. Gross Family Professor, Humphrey School of Public Affairs, UMN 2017- pres. Director, Master of Public Policy Program, Humphrey School of Public Affairs, UMN 2017- pres. Founding Director, Institute for Urban & Regional Infrastructure Finance, UMN 2012–2019 Associate Professor, Humphrey School of Public Affairs, UMN 2007–2011 Assistant Professor, Humphrey School of Public Affairs, UMN 2005–2007 Director, Political Science Internship Program, Eastern Michigan University 2005–2007 Assistant Professor, Department of Political Science, Eastern Michigan University Professional Experience 2002–2005 Education Program Specialist, Carl Vinson Institute of Government, UGA 1993–1998 Assistant Urban Planner, Xiamen Academy of Urban Planning & Design, China Awards and Honors 2020 SCPA Leadership Award, American Society for Public Administration (ASPA) 2018 Outstanding Services Award, Center for Transportation Studies, UMN 2018 Leadership Award, China-America -
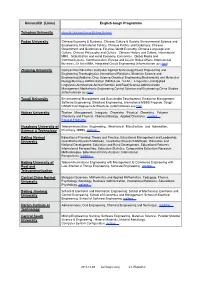
Universität (Links) English-Tough Programme Tsinghua University Fudan University Zhejiang University Tongji University Wuhan U
Universität (Links) English-tough Programme Tsinghua University aktuelle Informationen klicken Sie hier Fudan University Chinese Economy & Business, Chinese Culture & Society, Environmental Science and Engineering, International Politics, Chinese Politics and Diplomacy, Chinese Government and Governance, Finance, World Economy, Chinese Language and Culture, Chinese Philosophy and Culture, Chinese History and Culture, International MBA, Globalization and world Economy, Economics , Global Media and Communications, Communication, Europe and Asia in Global Affairs, International Business, S3 Asia MBA, Integrated Circuit Engineering (Informationen aus hier) Zhejiang University Comparative Education,Computer Applied Technology,Power Engineering and Engineering Thermophysics,International Relations, Materials Science and Engineering,Medicine, Crop Science,Chemical Engineering,Biochemistry and Molecular Biology,Business Administration (MBA),Law( LL.M.) ,Linguistics and Applied Linguistics,Architecture,Animal Nutrition and Feed Science,Administration Management,Mechatronic Engineering,Control Science and Engineering,China Studies (Informationen aus hier) Tongji University Environmental Management and Sustainable Development, Enterprise Management, Software Engineering, Structural Engineering, International MBBS Program, Tongji- UNSW Dual Degree in Architecture (Informationen aus hier) Wuhan University Tourism Management, Inorganic Chemistry, Physical Chemistry, Polymer Chemistry and Physical, Chemical Biology, Applied Chemistry , weitere…" Popular -

A Legacy in Chinese Education History, Or a Solution for Modern Undergraduates in China?
Journal of Education and Learning; Vol. 9, No. 6; 2020 ISSN 1927-5250 E-ISSN 1927-5269 Published by Canadian Center of Science and Education Chinese Shuyuan: A Legacy in Chinese Education History, or a Solution for Modern Undergraduates in China? Zhen Zeng1 1 School of Foreign Studies, Guangxi Normal University, Guilin, Guangxi, China Correspondence: Zhen Zeng, 82 Liuhe Road, Qixing District, Guilin, Guangxi, Postal Code: 541004, China. E-mail: [email protected] Received: September 28, 2020 Accepted: November 19, 2020 Online Published: November 26, 2020 doi:10.5539/jel.v9n6p173 URL: https://doi.org/10.5539/jel.v9n6p173 Abstract The paper looked into concepts claimed to be essence of Chinese residential college, an on-going institution presumed to be a solution towards undergraduates’ issues in some pioneer universities in China. It’s analyzed that Chinese residential college today in China is not a Shuyuan that was ever striving as a unique education mode in ancient China, even if it’s named after Shuyuan in Chinese, concerning on its nature, function and goal, while it’s not a conventional residential college in English speaking countries neither. By investigation and comparison of its origin, function and features among Shuyuan and Chinese residential college, the spirit of development of a human with goodness and well-being through pursuit of knowledge and culture inherited and transmitted in Shuyuan is unearthed, which is supposed to be the resource of inspiration when the pioneer universities and educators designed and operate residential college on Chinese campus, though the effects couldn’t be accounted as appealing as what Shuyuan produced in ancient China. -

Master in Law, Tongji University · 1988 - 1992: Bachelor in Law, Fudan University
WANG Zhenyu Associate Professor Department: Public Administration Email: [email protected] Office Phone: 021-65982272 EDUCATION BACKGROUND · 2004-2008: Ph.D. in Management, Tongji University · 1993-1996: Master in Law, Tongji University · 1988 - 1992: Bachelor in Law, Fudan University RESEARCH FIELD · Intellectual Property Management, Intellectual Property Policy WORK EXPERIENCE Teaching Experience · 2010-Now: Associate Professor, School of Economics and Management, Tongji University · 2006 - 2010: Lecturer, School of Economics and Management, Tongji University · 1999 - 2006: Lecturer, School of Literature and Law, Tongji University · 1996 - 1999: Assistant Professor, School of Literature and Law, Tongji University International Experience · 2013/08 - 2014/07: Visiting Scholar, sponsored by China Scholarship Council as the core member teacher, Drake University · 2007/08 - 2007/12: Visiting Scholar, intellectual property high-level talent selected by Shanghai Intellectual Property Administration and American International Education Foundation, Chicago-Kent of Law RESEARCH Sponsored Research Projects · “Intellectual Property rules in TPP and development strategies in China”, National Social Science Fund Project, 2016/07-2019/07 · “Intellectual Property public policy facing the challenges of TRIPS Agreement”, National Social Science Fund Project, 2011/07-2014/10 · “Cooperative Research on Chinese Intellectual Property Public Policy and TRIPs Agreement”, Shanghai Social Science Fund Project, 2010/06-2013/07 · “Intellectual Property Management -

(Gupes) Global Universities Partnership On
GUPES, 2013 GUPES, 2012 Morocco Shanghai, China GLOBALGLOBAL UNIVERSITIESUNIVERSITIES PARTNERSHIPPARTNERSHIP ONON ENVIRONMENTENVIRONMENT ANDAND SUSTAINABILITYSUSTAINABILITY (GUPES)(GUPES) OFFICIAL LAUNCH For more information on how to join GUPES, contact: Environmental Education and Training Unit Division of Environmental Policy Implementation United Nations Environment Programme P.O. Box 30552 00100 Nairobi, Kenya Tel: +254-20-7624101; Fax: +254-20-7623917 E-mail: [email protected] Website: unep.org/training UNEP-Tongji Institute of Envionment for Sustainable Development Tongji University 1239 Siping Road,Shanghai,China P.O. Box 200092 Tel: +86-21-65987790; Fax: +86-21-65987790 E-mail: [email protected] Website: http://unep-iesd.tongji.edu.cn 5-6 June 2012 Tongji University, Shanghai, China CONGRATULATION: The Official Lunch of Global Universities Part- nership on Environment and Sustainability Was Held Successfully at Tongji University! Prof. Wu Jiang as the Chair of the GUPES Offi- cial Steering Committee, as well as IESD as a University wide platform on sustainability issues! Preamble GUPES was the result of a consultative forum organized by UNEP and its partners to increase successful engagement with universities. It builds on the successes of the Mainstreaming Environment and Sustainability in A frican Universities (MESA), the nascent Mainstreaming Environment and Sustainability in the Caribbean Univer sities (MESCA) and the Asia - Pacific Regional University Consortium (RUC) . GUPES serves to increase the mainstreaming of environment and sustainability practices and curricula into universities around the world. It is also geared towards encouraging further interaction between UNEP and universities, a round the three pillars of education, training and applied research. This is done in accordance to the UN Decade of Education for Sustainable Development, and in partnership with UNESCO, UNU and other organizations. -

Ninghua, ZHONG (钟宁桦)
Ninghua, ZHONG (钟宁桦) Professor, Economics and Finance School of Economics and Management Tongji University, Shanghai [email protected]; [email protected] Academic Background PhD in Finance, Hong Kong University of Science and Technology, January 2013 PhD Committee: Sudipto Dasgupta, Vidan K. Goyal, K. C. John Wei, Albert Park, Zhigang Tao MA in Economics, Peking University, China Center for Economic Research, July 2008 Adviser: Yang Yao BA in Economics with highest distinction, Fudan University, School of Economics, July 2005 GPA: 3.82 out of 4.00 Class rank: 1 out of 109 Current Academic Appointment Professor, School of Economics and Management, Tongji University Promoted to Full Professor in December 2015 due to my exceptional performance Promoted to Associate Professor in June 2013 shortly after joining the university in March 2013 Teaches at one of the premier Executive MBA programs in the world (Tongji University/ENPC), ranked 68th in 2014 by the Financial Times Research interests include (a) applied microeconomics studies in the fields of labor economics, corporate finance, and empirical asset pricing and (b) Chinese economic reform and development Ningua, ZHONG (钟宁桦) 1 Publications and Working Papers—English As of December 2015, Google Scholar calculated 225 citations of my papers: On China’s Labor Market Issues (Papers in this field are developed from my master thesis) “Unions and Workers’ Welfare in Chinese Firms” (with Yang Yao), [download from JSTOR] Journal of Labor Economics 31, no. 3 (2013): 633–67. -
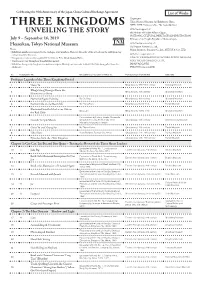
Three Kingdoms Unveiling the Story: List of Works
Celebrating the 40th Anniversary of the Japan-China Cultural Exchange Agreement List of Works Organizers: Tokyo National Museum, Art Exhibitions China, NHK, NHK Promotions Inc., The Asahi Shimbun With the Support of: the Ministry of Foreign Affairs of Japan, NATIONAL CULTURAL HERITAGE ADMINISTRATION, July 9 – September 16, 2019 Embassy of the People’s Republic of China in Japan With the Sponsorship of: Heiseikan, Tokyo National Museum Dai Nippon Printing Co., Ltd., Notes Mitsui Sumitomo Insurance Co.,Ltd., MITSUI & CO., LTD. ・Exhibition numbers correspond to the catalogue entry numbers. However, the order of the artworks in the exhibition may not necessarily be the same. With the cooperation of: ・Designation is indicated by a symbol ☆ for Chinese First Grade Cultural Relic. IIDA CITY KAWAMOTO KIHACHIRO PUPPET MUSEUM, ・Works are on view throughout the exhibition period. KOEI TECMO GAMES CO., LTD., ・ Exhibition lineup may change as circumstances require. Missing numbers refer to works that have been pulled from the JAPAN AIRLINES, exhibition. HIKARI Production LTD. No. Designation Title Excavation year / Location or Artist, etc. Period and date of production Ownership Prologue: Legends of the Three Kingdoms Period 1 Guan Yu Ming dynasty, 15th–16th century Xinxiang Museum Zhuge Liang Emerges From the 2 Ming dynasty, 15th century Shanghai Museum Mountains to Serve 3 Narrative Figure Painting By Qiu Ying Ming dynasty, 16th century Shanghai Museum 4 Former Ode on the Red Cliffs By Zhang Ruitu Ming dynasty, dated 1626 Tianjin Museum Illustrated -

Denver Seminar #2
Volume 62 #2 NATIONAL SOCIAL SCIENCE PROCEEDINGS Volume 62 #2 Denver, Colorado Summer Seminar 2016 Table of Contents Tourism and Liuzhou City’s Culture Based Performance Art: A Graphic Report Tian’an Lv, Central Washington University 1 Reform of Urban Underground Pipeline System Management in Liuzhou Guoliang Meng, Central Washington University 15 A Critical Look at the Charter School Debate Andrew Peiser, Mercy College, New York 25 Private-Public Partnerships (PPPS) and Industrial Park Construction in Liubei District Liang Qiong, Central Washington Uniserity 38 Cuba and the American Imagination: Shackles of the Past Umeme Sababu, Edinboro University of Pennsylvania 51 Speaking Without Tongues: Toward a More Humane Construct of the Online Learning Environment Ralph Lamar Turner, Carol Gassaway, Eastern Kentucky University 84 Taft’s Bathtub: Why One Size Does Not Fit All in the Comprehension and Application of Tinker, Bethel, Hazelwood, and Morse (Bong Hits) for Online School Law Classes for Administrator Candidates Charles R. Waggoner, Eastern New Mexico University 94 Contemporary Anticorruption Struggle in China Qiwen Wang, Central Washington University 101 The Liuzhou Municipal Bureau of Statistics (LMBS): The Evolution and Future of Statistical Reporting in China Dongyin Wei, Central Washington University 115 Student Athletes’ Definitions: Academic Integrity Lowell Wightman, Colorado State University 125 People’s Democracy in Luizhou City, China: Oversight, Transparency and Technology Jia Zhijuan, Yufeng District Legal Commission and Central Washington University Yu Zhou, Liuzhou Financial Bureau and Central Washington University Wu Nan, Liunan Sub-district Office and Central Washington University 138 Tourism and Liuzhou City’s Culture Based Performance Art: A Graphic Report Tian’an Lv Central Washington University 1 Introduction Cultural fusion is the defining characteristic of Liuzhou City’s emerging performance art tourism.