Biol. Pharm. Bull. 27(5) 661—669 (2004) 661
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Challenges and Countermeasures of Tourism
International Conference on Social Science and Technology Education (ICSSTE 2015) Challenges and Countermeasures of Regional Tourism Cooperation Development Strategy of Sichuan-Shanxi-Gansu Golden Triangle Area,Western China Qin Jianxiong1 Zhang Minmin1 1 College of tourism and historic culture, Southwest University For Natianalities, Chengdu, 610041 Abstract visitors can explore in this line up and down five SSGGTA triangle of three provinces , dependent thousand years of culture, enjoy the mystery of Qinba [1] landscape, folk customs are similar, for the first time landscape . These tourism resources in Chongqing, since the 2002 held in Bazhong of SSGGTA triangle area Chengdu, Xi'an, Lanzhou, Wuhan five source among SSGGTA triangle tourism cooperation zone is composed tourism cooperation will be signed in SSGGTA triangle of Sichuan Bazhong, Guangyuan, Dazhou and Shanxi tourism, build "Golden Triangle" cooperation agreement, Hanzhoung, Ankang three provinces and five to 2005 has successively held 3 annual meeting. The goal municipalities, carry out cooperation in the past 3 years, of cooperation is through the sincere cooperation of the three provinces and five municipalities in the propaganda, three provinces, the formation of regional tourism build mutual interaction, line group, strategic planning collaboration regular contact system, the characteristics of consensus interaction and so on has made significant tourism products, the formation of regional joint progress, regional cooperation has been fully affirmed the promotion,a barrier free Tourism Zone, to realize the two provincial government and support. Sichuan North Sichuan area has been the focus of tourism development sustainable development of Shanxi tourism in Golden in the province, tourism development, Shanxi will also Triangle. -

Sichuan Q I N G H a I G a N S U Christian Percentage of County/City Ruo'ergai
Sichuan Q i n g h a i G a n s u Christian Percentage of County/City Ruo'ergai Shiqu Jiuzhaigou S h a a n x i Hongyuan Aba Songpan Chaotian Qingchuan Nanjiang Seda Pingwu Lizhou Rangtang Wangcang Dege Heishui Zhaohua Tongjiang Ma'erkang Ganzi Beichuan Jiangyou Cangxi Wanyuan Mao Jiange Bazhou Enyang Zitong Pingchang Luhuo Jinchuan Li Anzhou Youxian Langzhong Xuanhan Mianzhu Yilong Shifang Fucheng Tongchuan Baiyu Luojiang Nanbu Pengzhou Yangting Xiaojin Jingyang Santai Yingshan Dachuan Danba Dujiangyan Xichong Xinlong Wenchuan Guanghan Peng'an Kaijiang Daofu Shehong Shunqing Qu Pi Xindu Zhongjiang Gaoping Chongzhou Jialing DayiWenjiang Jintang Pengxi Guang'an Dazhu Lushan Daying Yuechi Qianfeng Shuangliu Chuanshan Baoxing Qionglai Huaying T i b e t Batang Xinjin Jianyang Anju Wusheng Pujiang Kangding Pengshan Lezhi Linshui Mingshan Yanjiang Tianquan DanlengDongpo H u b e i Litang Yajiang Yucheng Renshou Anyue Yingjing Qingshen Zizhong Luding Jiajiang Jingyan Hongya Shizhong Weiyuan Dongxing Hanyuan Emeishan Rong Shizhong WutongqiaoGongjing Da'an Longchang C h o n g q i n g Xiangcheng Shimian Jinkouhe Shawan Ziliujing Yantan Ebian Qianwei Lu Jiulong Muchuan Fushun Daocheng Ganluo Longmatan Derong Xuzhou NanxiJiangyang Mabian Pingshan Cuiping Hejiang Percent Christian Naxi Mianning Yuexi Jiang'an Meigu Changning (County/City) Muli Leibo Gao Gong Xide Xingwen 0.8% - 3% Zhaojue Junlian Xuyong Gulin Chengdu area enlarged 3.1% - 4% Xichang Jinyang Qingbaijiang Yanyuan Butuo Pi Puge Xindu 4.1% - 5% Dechang Wenjiang Y u n n a n Jinniu Chenghua Qingyang 5.1% - 6% Yanbian Ningnan Miyi G u i z h o u Wuhou Longquanyi 6.1% - 8.8% Renhe Jinjiang Xi Dong Huidong Shuangliu Renhe Huili Disputed boundary with India Data from Asia Harvest, www.asiaharvest.org. -

Study on the Ecotourism Development in Dazhou
Open Journal of Social Sciences, 2018, 6, 24-34 http://www.scirp.org/journal/jss ISSN Online: 2327-5960 ISSN Print: 2327-5952 Study on the Ecotourism Development in Dazhou Xiaomei Pu1, Lin Tian2, Zibiao Cheng3 1Research Center of Sichuan Old Revolutionary Areas Development, Sichuan University of Arts and Science, Dazhou, China 2School of Foreign Languages, Sichuan University of Arts and Science, Dazhou, China 3School of Finance and Economics Management, Sichuan University of Arts and Science, Dazhou, China How to cite this paper: Pu, X.M., Tian, L. Abstract and Cheng, Z.B. (2018) Study on the Eco- tourism Development in Dazhou. Open After comprehensive discussion of the origin of ecotourism, the concept of Journal of Social Sciences, 6, 24-34. ecotourism and the theoretical basis for ecotourism development, the paper https://doi.org/10.4236/jss.2018.65002 carried out the SWOT analysis on ecotourism development in Dazhou City, Received: April 8, 2018 and then proposed development strategies. The strategies were to: enhance Accepted: May 13, 2018 the ecological awareness of the entire people and create a good atmosphere for Published: May 16, 2018 ecotourism development; break the talent bottleneck of ecotourism develop- ment by adopting the policy of “combination boxing”; make scientific and Copyright © 2018 by authors and Scientific Research Publishing Inc. feasible master plan for Dazhou’s ecotourism development; develop quality This work is licensed under the Creative ecotourism products; innovate marketing strategies for ecotourism in Dazhou. Commons Attribution International License (CC BY 4.0). Keywords http://creativecommons.org/licenses/by/4.0/ Open Access Dazhou, Ecotourism, Development 1. -

Qinghai WLAN Area 1/13
Qinghai WLAN area NO. SSID Location_Name Location_Type Location_Address City Province 1 ChinaNet Quality Supervision Mansion Business Building No.31 Xiguan Street Xining City Qinghai Province No.160 Yellow River Road 2 ChinaNet Victory Hotel Conference Center Convention Center Xining City Qinghai Province 3 ChinaNet Shangpin Space Recreation Bar No.16-36 Xiguan Street Xining City Qinghai Province 4 ChinaNet Business Building No.372 Qilian Road Xining City Qinghai Province Salt Mansion 5 ChinaNet Yatai Trade City Large Shopping Mall Dongguan Street Xining City Qinghai Province 6 ChinaNet Gome Large Shopping Mall No.72 Dongguan Street Xining City Qinghai Province 7 ChinaNet West Airport Office Building Business Building No.32 Bayi Road Xining City Qinghai Province Government Agencies 8 ChinaNet Chengdong District Government Xining City Qinghai Province and Other Institutions Delingha Road 9 ChinaNet Junjiao Mansion Business Building Xining City Qinghai Province Bayi Road Government Agencies 10 ChinaNet Higher Procuratortate Office Building Xining City Qinghai Province and Other Institutions Wusi West Road 11 ChinaNet Zijin Garden Business Building No.41, Wusi West Road Xining City Qinghai Province 12 ChinaNet Qingbai Shopping Mall Large Shopping Mall Xining City Qinghai Province No.39, Wusi Avenue 13 ChinaNet CYTS Mansion Business Building No.55-1 Shengli Road Xining City Qinghai Province 14 ChinaNet Chenxiong Mansion Business Building No.15 Shengli Road Xining City Qinghai Province 15 ChinaNet Platform Bridge Shoes City Large Shopping -

Table of Codes for Each Court of Each Level
Table of Codes for Each Court of Each Level Corresponding Type Chinese Court Region Court Name Administrative Name Code Code Area Supreme People’s Court 最高人民法院 最高法 Higher People's Court of 北京市高级人民 Beijing 京 110000 1 Beijing Municipality 法院 Municipality No. 1 Intermediate People's 北京市第一中级 京 01 2 Court of Beijing Municipality 人民法院 Shijingshan Shijingshan District People’s 北京市石景山区 京 0107 110107 District of Beijing 1 Court of Beijing Municipality 人民法院 Municipality Haidian District of Haidian District People’s 北京市海淀区人 京 0108 110108 Beijing 1 Court of Beijing Municipality 民法院 Municipality Mentougou Mentougou District People’s 北京市门头沟区 京 0109 110109 District of Beijing 1 Court of Beijing Municipality 人民法院 Municipality Changping Changping District People’s 北京市昌平区人 京 0114 110114 District of Beijing 1 Court of Beijing Municipality 民法院 Municipality Yanqing County People’s 延庆县人民法院 京 0229 110229 Yanqing County 1 Court No. 2 Intermediate People's 北京市第二中级 京 02 2 Court of Beijing Municipality 人民法院 Dongcheng Dongcheng District People’s 北京市东城区人 京 0101 110101 District of Beijing 1 Court of Beijing Municipality 民法院 Municipality Xicheng District Xicheng District People’s 北京市西城区人 京 0102 110102 of Beijing 1 Court of Beijing Municipality 民法院 Municipality Fengtai District of Fengtai District People’s 北京市丰台区人 京 0106 110106 Beijing 1 Court of Beijing Municipality 民法院 Municipality 1 Fangshan District Fangshan District People’s 北京市房山区人 京 0111 110111 of Beijing 1 Court of Beijing Municipality 民法院 Municipality Daxing District of Daxing District People’s 北京市大兴区人 京 0115 -

China's “Bilingual Education” Policy in Tibet Tibetan-Medium Schooling Under Threat
HUMAN CHINA’S “BILINGUAL EDUCATION” RIGHTS POLICY IN TIBET WATCH Tibetan-Medium Schooling Under Threat China's “Bilingual Education” Policy in Tibet Tibetan-Medium Schooling Under Threat Copyright © 2020 Human Rights Watch All rights reserved. Printed in the United States of America ISBN: 978-1-6231-38141 Cover design by Rafael Jimenez Human Rights Watch defends the rights of people worldwide. We scrupulously investigate abuses, expose the facts widely, and pressure those with power to respect rights and secure justice. Human Rights Watch is an independent, international organization that works as part of a vibrant movement to uphold human dignity and advance the cause of human rights for all. Human Rights Watch is an international organization with staff in more than 40 countries, and offices in Amsterdam, Beirut, Berlin, Brussels, Chicago, Geneva, Goma, Johannesburg, London, Los Angeles, Moscow, Nairobi, New York, Paris, San Francisco, Sydney, Tokyo, Toronto, Tunis, Washington DC, and Zurich. For more information, please visit our website: http://www.hrw.org MARCH 2020 ISBN: 978-1-6231-38141 China's “Bilingual Education” Policy in Tibet Tibetan-Medium Schooling Under Threat Map ........................................................................................................................ i Summary ................................................................................................................ 1 Chinese-Medium Instruction in Primary Schools and Kindergartens .......................................... 2 Pressures -

Save the Children in China 2013 Annual Review
Save the Children in China 2013 Annual Review Save the Children in China 2013 Annual Review i CONTENTS 405,579 In 2013, Save the Children’s child education 02 2013 for Save the Children in China work helped 405,579 children and 206,770 adults in China. 04 With Children and For Children 06 Saving Children’s Lives 08 Education and Development 14 Child Protection 16 Disaster Risk Reduction and Humanitarian Relief 18 Our Voice for Children 1 20 Media and Public Engagement 22 Our Supporters Save the Children organised health and hygiene awareness raising activities in the Nagchu Prefecture of Tibet on October 15th, 2013 – otherwise known as International Handwashing Day. In addition to teaching community members and elementary school students how to wash their hands properly, we distributed 4,400 hygiene products, including washbasins, soap, toothbrushes, toothpastes, nail clippers and towels. 92,150 24 Finances In 2013, we responded to three natural disasters in China, our disaster risk reduction work and emergency response helped 92,150 Save the Children is the world’s leading independent children and 158,306 adults. organisation for children Our vision A world in which every child attains the right to survival, protection, development and 48,843 participation In 2013, our child protection work in China helped 48,843 children and 75,853 adults. Our mission To inspire breakthroughs in the way the world treats children, and to achieve immediate and 2 lasting change in their lives Our values 1 Volunteers cheer on Save the Children’s team at the Beijing Marathon on October 20th 2013. -

China PROJECT DOCUMENT
United Nations Development Programme Country: China PROJECT DOCUMENT Project Title: Strengthening the effectiveness of the protected area system in Qinghai Province, China to conserve globally important biodiversity UNDAF Outcome(s): Outcome 1.2. Policy and implementation mechanisms to manage natural resources are strengthened, with special attention to poor and vulnerable groups UNDP Strategic Plan Environment and Sustainable Development Primary Outcome: Mobilizing environmental financing UNDP Strategic Plan Secondary Outcome: Mainstreaming environment and energy Expected CP Outcome(s ): Low carbon and other environmentally sustainable strategies and technologies are adopted widely to meet China’s commitments and compliance with Multilateral Environmental Agreements. Provincial capacities of key institutions are strengthened to implement global environmental commitments at regional level through integration of biodiversity and other environmental concerns into regional policies and programmes involved. Expected CPAP Output(s): Capacity to implement local climate change action plans for mitigation and adaptation, and sustainable development built. Executing Entity/Implementing Partner: Department of Forestry, Qinghai Province Government, China Implementing Entity/Responsible Partners: Ministry of Environmental Protection (through umbrella project China Biodiversity Partnership and Framework for Action) UNDP GEF PIMS 4179 GEF Project ID 3992 Brief description As the fourth largest province in China, with a total area of 720,000 km 2, Qinghai serves as a significant store of the national biodiversity, exhibits some unique high altitude grassland, mountain, wetland, desert and forest ecosystems, and serves as a significant controller of the Asian monsoon system that affects the climate of 3 billion people. The province includes the headwaters of three of Asia’s major rivers – the Yellow, Yangtze and Mekong rivers. -

In China 2014 Annual Review
Save the Children in China 2014 Annual Review Save the Children in China 2014 Annual Review i 2014 · Snapshot CONTENTS 02 Stories for 2014 04 In the world and in China 12 06 Saving Children’s Lives In 2014, Save the Children worked in Education 12 provinces (autonomous regions and 08 municipalities) in Mainland China, including Child Protection Shaanxi and Jiangsu provinces for the first time. 14 16 Disaster Risk Reduction and Humanitarian Relief 18 Our Voice for Children 1 1.09 MILLION 20 Media and Campaigns In 2014, Save the Children helped 1,090,752 children and 1,546,826 adults in China. 22 Our Supporters In November 2014, a mother brought her child to see the doctor in the village clinic in Qigelike Village, Sayibage Township, Moyu County, Xinjiang. Save the Children implemented the "Integrated Management of Childhood Illnesses" Project in Moyu County in order to build the capacity of grassroots health workers in diagnosing and treating common childhood diseases. Photo credit: Nurmamat Nurjan 24 Finances MILLION 10 Save the Children is the world’s leading independent In 2014, our media and public campaign work organisation for children reached an audience of more than 10 million. Our vision 2 A world in which every child attains the right to survival, protection, development and participation Our mission 75% To inspire breakthroughs in the way the world treats children, and to achieve immediate and Cover A girl in the ECCD centre in Mojiang County, Yunnan Province. Photo credit: Liu Chunhua 1 In June 2014, Yumiao Elementary School, a private school in Shanghai, organised family-school cooperation activities. -

Chem. Pharm. Bull. 54(11) 1491—1499 (2006) 1491
November 2006 Chem. Pharm. Bull. 54(11) 1491—1499 (2006) 1491 Comparative Study of Chemical Constituents of Rhubarb from Different Origins ,a,b a a c d Katsuko KOMATSU,* Yorinobu NAGAYAMA, Ken TANAKA, Yun LING, Shao-Qing CAI, a e Takayuki OMOTE, and Meselhy Ragab MESELHY a Division of Pharmacognosy, Department of Medicinal Resources, Institute of Natural Medicine, University of Toyama; b21st Century COE Program, University of Toyama; 2630 Sugitani, Toyama 930–0194, Japan: c Yanjing Hospital; Beijing 100083, People’s Republic of China: d Department of Natural Medicines, School of Pharmaceutical Sciences, Peking University; Beijing 100083, People’s Republic of China: and e Department of Pharmacognosy and Medicinal Plants, Faculty of Pharmacy, Cairo University; Kasr EL-Ainy, Cairo, Egypt. Received April 25, 2006; accepted July 24, 2006 A comparative study of the pharmacologically active constituents of 24 rhubarb samples, which were identi- fied genetically as Rheum tanguticum, 3 intraspecies groups of R. palmatum and R. officinale, was conducted using reversed-phase high performance liquid chromatography (HPLC) methods. Thirty compounds belonging to anthraquinones, anthraquinone glucosides, dianthrones, phenylbutanones, stilbenes, flavan-3-ols, procyani- dins, galloylglucoses, acylglucoses, gallic acid, and polymeric procyanidins were analyzed quantitatively. The drug samples derived from the same botanical source showed similar chromatographic profiles, and the compa- rable specific shape that appeared in the 10-directed radar graphs constructed on the basis of the results of quantitative analysis indicated the relationship between chemical constituent patterns and genetic varieties of rhubarb samples. Key words Rhei Rhizoma; Rheum; genetic variety; HPLC; quantitative comparison Rhei Rhizoma (rhubarb), called Dahuang in Chinese, is has been observed within the genera Panax,8) Glycyrrhiza,9) widely known as a purgative and anti-inflammatory agent. -

Taxonomy and Phylogenetic Relationship of Zokors
Journal of Genetics (2020)99:38 Ó Indian Academy of Sciences https://doi.org/10.1007/s12041-020-01200-2 (0123456789().,-volV)(0123456789().,-volV) RESEARCH ARTICLE Taxonomy and phylogenetic relationship of zokors YAO ZOU1, MIAO XU1, SHIEN REN1, NANNAN LIANG1, CHONGXUAN HAN1*, XIAONING NAN1* and JIANNING SHI2 1Key Laboratory of National Forestry and Grassland Administration on Management of Western Forest Bio-Disaster, Northwest Agriculture and Forestry University, Yangling 712100, People’s Republic of China 2Ningxia Hui Autonomous Region Forest Disease and Pest Control Quarantine Station, Yinchuan 750001, People’s Republic of China *For correspondence. E-mail: Chongxuan Han, [email protected]; Xiaoning Nan, [email protected]. Received 24 October 2019; revised 19 February 2020; accepted 2 March 2020 Abstract. Zokor (Myospalacinae) is one of the subterranean rodents, endemic to east Asia. Due to the convergent and parallel evolution induced by its special lifestyles, the controversies in morphological classification of zokor appeared at the level of family and genus. To resolve these controversies about taxonomy and phylogeny, the phylogenetic relationships of 20 species of Muroidea and six species of zokors were studied based on complete mitochondrial genome and mitochondrial Cytb gene, respectively. Phylogeny analysis of 20 species of Muroidea indicated that the zokor belonged to the family Spalacidae, and it was closer to mole rat rather than bamboo rat. Besides, by investigating the phylogenetic relationships of six species of zokors, the status of two genera of Eospalax and Myospalax was affirmed because the two clades differentiated in phylogenetic tree represented two types of zokors, convex occiput type and flat occiput type, respectively. -
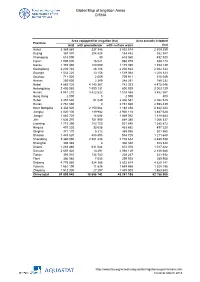
Global Map of Irrigation Areas CHINA
Global Map of Irrigation Areas CHINA Area equipped for irrigation (ha) Area actually irrigated Province total with groundwater with surface water (ha) Anhui 3 369 860 337 346 3 032 514 2 309 259 Beijing 367 870 204 428 163 442 352 387 Chongqing 618 090 30 618 060 432 520 Fujian 1 005 000 16 021 988 979 938 174 Gansu 1 355 480 180 090 1 175 390 1 153 139 Guangdong 2 230 740 28 106 2 202 634 2 042 344 Guangxi 1 532 220 13 156 1 519 064 1 208 323 Guizhou 711 920 2 009 709 911 515 049 Hainan 250 600 2 349 248 251 189 232 Hebei 4 885 720 4 143 367 742 353 4 475 046 Heilongjiang 2 400 060 1 599 131 800 929 2 003 129 Henan 4 941 210 3 422 622 1 518 588 3 862 567 Hong Kong 2 000 0 2 000 800 Hubei 2 457 630 51 049 2 406 581 2 082 525 Hunan 2 761 660 0 2 761 660 2 598 439 Inner Mongolia 3 332 520 2 150 064 1 182 456 2 842 223 Jiangsu 4 020 100 119 982 3 900 118 3 487 628 Jiangxi 1 883 720 14 688 1 869 032 1 818 684 Jilin 1 636 370 751 990 884 380 1 066 337 Liaoning 1 715 390 783 750 931 640 1 385 872 Ningxia 497 220 33 538 463 682 497 220 Qinghai 371 170 5 212 365 958 301 560 Shaanxi 1 443 620 488 895 954 725 1 211 648 Shandong 5 360 090 2 581 448 2 778 642 4 485 538 Shanghai 308 340 0 308 340 308 340 Shanxi 1 283 460 611 084 672 376 1 017 422 Sichuan 2 607 420 13 291 2 594 129 2 140 680 Tianjin 393 010 134 743 258 267 321 932 Tibet 306 980 7 055 299 925 289 908 Xinjiang 4 776 980 924 366 3 852 614 4 629 141 Yunnan 1 561 190 11 635 1 549 555 1 328 186 Zhejiang 1 512 300 27 297 1 485 003 1 463 653 China total 61 899 940 18 658 742 43 241 198 52