Modularity and Neural Coding from a Brainstem Synaptic Wiring Diagram
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Building Atlases of the Brain
© Copyright, Princeton University Press. No part of this book may be distributed, posted, or reproduced in any form by digital or mechanical means without prior written permission of the publisher. BUILDING AtLASES OF THE BRAIN Mike Hawrylycz With Chinh Dang, Christof Koch, and Hongkui Zeng A Very Brief History of Brain Atlases The earliest known significant works on human anatomy were collected by the Greek physician Claudius Galen around 200 BCE. This ancient corpus remained the dominant viewpoint through the Middle Ages until the classic work De humani corporis fabrica (On the Fabric of the Human Body) by Andreas Vesalius of Padua (1514–1564), the first mod- ern anatomist. Even today many of Vesalius’s drawings are astonishing to study and are largely accurate. For nearly two centuries scholars have recognized that the brain is compartmentalized into distinct regions, and this organization is preserved throughout mammals in general. However, comprehending the structural organization and function of the nervous system remains one of the primary challenges in neuro- science. To analyze and record their findings neuroanatomists develop atlases or maps of the brain similar to those cartographers produce. The state of our understanding today of an integrated plan of brain function remains incomplete. Rather than indicating a lack of effort, this observation highlights the profound complexity and interconnec- tivity of all but the simplest neural structures. Laying the foundation of cellular neuroscience, Santiago Ramón y Cajal (1852– 1934) drew and classified many types of neurons and speculated that the brain consists of an interconnected network of distinct neurons, as opposed to a more continuous web. -

Spiking Allows Neurons to Estimate Their Causal Effect
bioRxiv preprint doi: https://doi.org/10.1101/253351; this version posted January 25, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Spiking allows neurons to estimate their causal effect Benjamin James Lansdell1,+ and Konrad Paul Kording1 1Department of Bioengineering, University of Pennsylvania, PA, USA [email protected] Learning aims at causing better performance and the typical gradient descent learning is an approximate causal estimator. However real neurons spike, making their gradients undefined. Interestingly, a popular technique in economics, regression discontinuity design, estimates causal effects using such discontinuities. Here we show how the spiking threshold can reveal the influence of a neuron's activity on the performance, indicating a deep link between simple learning rules and economics-style causal inference. Learning is typically conceptualized as changing a neuron's properties to cause better performance or improve the reward R. This is a problem of causality: to learn, a neuron needs to estimate its causal influence on reward, βi. The typical solution linearizes the problem and leads to popular gradient descent- based (GD) approaches of the form βGD = @R . However gradient descent is just one possible approximation i @hi to the estimation of causal influences. Focusing on the underlying causality problem promises new ways of understanding learning. Gradient descent is problematic as a model for biological learning for two reasons. First, real neurons spike, as opposed to units in artificial neural networks (ANNs), and their rates are usually quite slow so that the discreteness of their output matters (e.g. -
Glia and Neurons Glia and Neurons Objective: Role of Glia in Nervous System Agenda: 1
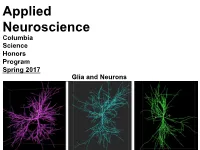
Applied Neuroscience Columbia Science Honors Program Spring 2017 Glia and Neurons Glia and Neurons Objective: Role of Glia in Nervous System Agenda: 1. Glia • Guest Lecture by Dr. Jennifer Ziegenfuss 2. Machine Learning • Applications in Neuroscience Connectomics A connectome is a comprehensive map of neural connections in the brain (wiring diagram). • Fails to illustrate how neurons behave in real-time (neural dynamics) • Fails to show how a behavior is generated • Fails to account for glia A Tiny Piece of the Connectome Serial EM Reconstruction of Axonal Inputs (various colors) onto a section of apical dendrite (grey) of a pyramidal neuron in mouse cerebral cortex. Arrows mark functional synapses. Lichtman Lab (Harvard) Scaling up efforts to map neural connections is unlikely to deliver the promised benefits such as understanding: • how the brain produces memories • Perception • Consciousness • Treatments for epilepsy, depression, and schizophrenia While glia have been neglected in the quest to understand neuronal signaling, they can sense neuronal activity and control it. Are Glia the Genius Cells? Structure-Function Divide in Brain The function of a neural network is critically dependent upon its interconnections. Only C. elegans has a complete connectome. In neuroscience: • many common diseases and disorders have no known histological trace • debates how many cell types there are • questionable plan to image whole volumes by EM • complexity of structure How is the brain’s function related to its complex structure? Worm Connectome Dots are individual neurons and lines represent axons. C. elegans Caenorhabditis elegans (C. elegans) is a transparent nematode commonly used in neuroscience research. They have a simple nervous system: 302 neurons and 7000 synapses. -

UNDERSTANDING the BRAIN Tbook Collections
FROM THE NEW YORK TIMES ARCHIVES UNDERSTANDING THE BRAIN TBook Collections Copyright © 2015 The New York Times Company. All rights reserved. Cover Photograph by Zach Wise for The New York Times This ebook was created using Vook. All of the articles in this work originally appeared in The New York Times. eISBN: 9781508000877 The New York Times Company New York, NY www.nytimes.com www.nytimes.com/tbooks Obama Seeking to Boost Study of Human Brain By JOHN MARKOFF FEB. 17, 2013 The Obama administration is planning a decade-long scientific effort to examine the workings of the human brain and build a comprehensive map of its activity, seeking to do for the brain what the Human Genome Project did for genetics. The project, which the administration has been looking to unveil as early as March, will include federal agencies, private foundations and teams of neuroscientists and nanoscientists in a concerted effort to advance the knowledge of the brain’s billions of neurons and gain greater insights into perception, actions and, ultimately, consciousness. Scientists with the highest hopes for the project also see it as a way to develop the technology essential to understanding diseases like Alzheimer’sand Parkinson’s, as well as to find new therapies for a variety of mental illnesses. Moreover, the project holds the potential of paving the way for advances in artificial intelligence. The project, which could ultimately cost billions of dollars, is expected to be part of the president’s budget proposal next month. And, four scientists and representatives of research institutions said they had participated in planning for what is being called the Brain Activity Map project. -

Application of Stochastic Modeling Techniques to Long-Term, High-Resolution Time-Lapse Microscopy Of
Quantitative analysis of axon elongation: A novel application of stochastic modeling techniques to long-term, high-resolution time-lapse microscopy of cortical axons MASSACHUSETTS INSTTilTE OF TECHNOLOGY by APR 0 1 2010 Neville Espi Sanjana LIBRARIES B.S., Symbolic Systems, Stanford University (2001) A.B., English, Stanford University (2001) Submitted to the Department of Brain & Cognitive Sciences in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Neuroscience at the MASSACHUSETTS INSTITUTE OF TECHNOLOGY February 2010 @ Massachusetts Institute of Technology 2010. All rights reserved. A u th or ....... ...................................... Department of Brain & Cognitive Sciences December 17, 2009 Certified by..... H. Sebastian Seung Professor of Computational Neuroscience - 40" / Thesis Supervisor Accepted by............................ Earl . Miller Earl K. Miller Picower Professor of Neuroscience Chairman, Department Committee on Graduate Theses Quantitative analysis of axon elongation: A novel application of stochastic modeling techniques to long-term, high-resolution time-lapse microscopy of cortical axons by Neville Espi Sanjana Submitted to the Department of Brain & Cognitive Sciences on December 17, 2009, in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Neuroscience Abstract Axons exhibit a rich variety of behaviors, such as elongation, turning, branching, and fas- ciculation, all in service of the complex goal of wiring up the brain. In order to quantify these behaviors, I have developed a system for in vitro imaging of axon growth cones with time-lapse fluorescence microscopy. Image tiles are automatically captured and assembled into a mosaic image of a square millimeter region. GFP-expressing mouse cortical neurons can be imaged once every few minutes for up to weeks if phototoxicity is minimized. -

Design and Evaluation of Interactive Proofreading Tools for Connectomics
Design and Evaluation of Interactive Proofreading Tools for Connectomics Daniel Haehn, Seymour Knowles-Barley, Mike Roberts, Johanna Beyer, Narayanan Kasthuri, Jeff W. Lichtman, and Hanspeter Pfister Fig. 1: Proofreading with Dojo. We present a web-based application for interactive proofreading of automatic segmentations of connectome data acquired via electron microscopy. Split, merge and adjust functionality enables multiple users to correct the labeling of neurons in a collaborative fashion. Color-coded structures can be explored in 2D and 3D. Abstract—Proofreading refers to the manual correction of automatic segmentations of image data. In connectomics, electron mi- croscopy data is acquired at nanometer-scale resolution and results in very large image volumes of brain tissue that require fully automatic segmentation algorithms to identify cell boundaries. However, these algorithms require hundreds of corrections per cubic micron of tissue. Even though this task is time consuming, it is fairly easy for humans to perform corrections through splitting, merging, and adjusting segments during proofreading. In this paper we present the design and implementation of Mojo, a fully-featured single- user desktop application for proofreading, and Dojo, a multi-user web-based application for collaborative proofreading. We evaluate the accuracy and speed of Mojo, Dojo, and Raveler, a proofreading tool from Janelia Farm, through a quantitative user study. We designed a between-subjects experiment and asked non-experts to proofread neurons in a publicly available connectomics dataset. Our results show a significant improvement of corrections using web-based Dojo, when given the same amount of time. In addition, all participants using Dojo reported better usability. We discuss our findings and provide an analysis of requirements for designing visual proofreading software. -

An Error Detection and Correction Framework for Connectomics
An Error Detection and Correction Framework for Connectomics Jonathan Zung Ignacio Tartavull∗ Kisuk Leey Princeton University Princeton University MIT [email protected] [email protected] [email protected] H. Sebastian Seung Princeton University [email protected] Abstract We define and study error detection and correction tasks that are useful for 3D reconstruction of neurons from electron microscopic imagery, and for image seg- mentation more generally. Both tasks take as input the raw image and a binary mask representing a candidate object. For the error detection task, the desired output is a map of split and merge errors in the object. For the error correction task, the desired output is the true object. We call this object mask pruning, because the candidate object mask is assumed to be a superset of the true object. We train multiscale 3D convolutional networks to perform both tasks. We find that the error-detecting net can achieve high accuracy. The accuracy of the error-correcting net is enhanced if its input object mask is “advice” (union of erroneous objects) from the error-detecting net. 1 Introduction While neuronal circuits can be reconstructed from volumetric electron microscopic imagery, the process has historically [39] and even recently [37] been highly laborious. One of the most time- consuming reconstruction tasks is the tracing of the brain’s “wires,” or neuronal branches. This task is an example of instance segmentation, and can be automated through computer detection of the boundaries between neurons. Convolutional networks were first applied to neuronal boundary detection a decade ago [14, 38]. Since then convolutional nets have become the standard approach, arXiv:1708.02599v2 [cs.CV] 3 Dec 2017 and the accuracy of boundary detection has become impressively high [40, 3, 21, 9]. -

Seymour Knowles-Barley1*, Mike Roberts2, Narayanan Kasthuri1, Dongil Lee1, Hanspeter Pfister2 and Jeff W. Lichtman1
Mojo 2.0: Connectome Annotation Tool Seymour Knowles-Barley1*, Mike Roberts2, Narayanan Kasthuri1, Dongil Lee1, Hanspeter Pfister2 and Jeff W. Lichtman1 1Harvard University, Department of Molecular and Cellular Biology, USA 2Harvard University, School of Engineering and Applied Sciences, USA Available online at http://www.rhoana.org/ Introduction A connectome is the wiring diagram of connections in a nervous system. Mapping this network of connections is necessary for discovering the underlying architecture of the brain and investigating the physical underpinning of cognition, intelligence, and consciousness [1, 2, 3]. It is also an important step in understanding how connectivity patterns are altered by mental illnesses, learning disorders, and age related changes in the brain. Fully automatic computer vision techniques are available to segment electron microscopy (EM) connectome data. Currently we use the Rhoana pipeline to process images, but results are still far from perfect and require additional human annotation to produce an accurate connectivity map [4]. Here we present Mojo 2.0, an open source, interactive, scalable annotation tool to correct errors in automatic segmentation results. Mojo Features ● Interactive annotation tool ● Smart scribble interface for split and merge operations ● Scalable up to TB scale volumes ● Entire segmentation pipeline including The Mojo interface displaying an EM section from mouse cortex, approximately 7.5x5μm. Mojo is open source and available online: http://www.rhoana.org/ Annotation Results A mouse brain cortex dataset was annotated with the Rhoana image processing pipeline and corrected using Mojo. Partial annotations over two sub volumes totaling 2123μm3 were made by novice users. On average, 1μm3 was annotated in 15 minutes and required 126 edit operations. -

Publications of Sebastian Seung
Publications of Sebastian Seung V. Jain, H. S. Seung, and S. C. Turaga. Machines that learn to segment images: a crucial technology for connectomics. (pdf) Curr. Opin. Neurobiol (2010). V. Jain, B. Bollmann, M. Richardson, D. Berger et al. Boundary learning by optimization with topological constraints. (pdf supplementary info) Proceedings of the IEEE 23rd Conference on Computer Vision and Pattern Recognition (2010). I. R. Wickersham, H. A. Sullivan, and H. S. Seung. Production of glycoprotein-deleted rabies viruses for monosynaptic tracing and high-level gene expression in neurons. (pdf) Nature Protocols. 5, 596-606 (2010). S. C. Turaga, J. F. Murray, V. Jain, F. Roth, M. Helmstaedter, K. Briggman, W. Denk, and H. S. Seung. Convolutional networks can learn to generate affinity graphs for image segmentation. (pdf) Neural Computation 22, 511-538 (2010). S. C. Turaga, K. L. Briggman, M. Helmstaedter, W. Denk, and H. S. Seung. Maximin affinity learning of image segmentation. (pdf) Adv. Neural Info. Proc. Syst. 22 (Proceedings of NIPS '09) (2010). J. Wang, M. T. Hasan, and H. S. Seung. Laser-evoked synaptic transmission in cultured hippocampal neurons expressing Channelrhodopsin-2 delivered by adeno-associated virus.(pdf) J. Neurosci. Methods 183, 165-175 (2009). Y. Loewenstein, D. Prelec, and H. S. Seung. Operant matching as a Nash equilibrium of an intertemporal game. (pdf) Neural Computation 21, 2755-2773 (2009). H. S. Seung. Reading the Book of Memory: Sparse Sampling versus Dense Mapping of Connectomes. (pdf) Neuron 62, 17-29 (2009). V. Jain and H. S. Seung. Natural Image Denoising with Convolutional Networks (pdf) Adv. Neural Info. -

Massive Data Management and Sharing Module for Connectome Reconstruction
brain sciences Article Massive Data Management and Sharing Module for Connectome Reconstruction Jingbin Yuan 1 , Jing Zhang 1, Lijun Shen 2,*, Dandan Zhang 2, Wenhuan Yu 2 and Hua Han 3,4,5,* 1 School of Automation, Harbin University of Science and Technology, Harbin 150080, China; [email protected] (J.Y.); [email protected] (J.Z.) 2 Research Center for Brain-inspired Intelligence, Institute of Automation, Chinese Academy of Sciences, Beijing 100190, China; [email protected] (D.Z.); [email protected] (W.Y.) 3 The National Laboratory of Pattern Recognition, Institute of Automation, Chinese Academy of Sciences, Beijing 100190, China 4 Center for Excellence in Brain Science and Intelligence Technology, Chinese Academy of Sciences, Shanghai 200031, China 5 The School of Future Technology, University of Chinese Academy of Sciences, Beijing 100049, China * Correspondence: [email protected] (L.S.); [email protected] (H.H); Tel.: +86-010-82544385 (L.S.); +86-010-82544710 (H.H) Received: 30 April 2020; Accepted: 20 May 2020; Published: 22 May 2020 Abstract: Recently, with the rapid development of electron microscopy (EM) technology and the increasing demand of neuron circuit reconstruction, the scale of reconstruction data grows significantly. This brings many challenges, one of which is how to effectively manage large-scale data so that researchers can mine valuable information. For this purpose, we developed a data management module equipped with two parts, a storage and retrieval module on the server-side and an image cache module on the client-side. On the server-side, Hadoop and HBase are introduced to resolve massive data storage and retrieval. -

Connectome Seymour Knowles-Barley1,2, Thouis R
CATEGORY: COMPUTER VISION - CV04 POSTER CONTACT NAME P4212 Thouis Jones: [email protected] Deep Learning for the Connectome Seymour Knowles-Barley1,2, Thouis R. Jones1,2, Josh Morgan2, Dongil Lee2, Narayanan Kasthuri2, Jeff W. Lichtman2 and Hanspeter Pfister1. 1Harvard University, School of Engineering and Applied Sciences, USA. 2Harvard University, Department of Molecular and Cellular Biology, USA. Introduction The connectome is the wiring diagram of the nervous system. Mapping this network is necessary for discovering the underlying architecture of the brain and investigating the physical underpinning of cognition, intelligence, and consciousness [1, 2, 3]. It is also an important step in understanding how connectivity patterns are altered by mental illnesses, learning disorders, and age related changes in the brain. In our experiments, brain tissue is microtomed and imaged with an electron microscope at a resolution of 4x4x30nm per voxel. Cell membranes and sub-cellular structures such as mitochondria, synapses and vesicles are clearly visible in the images. We have developed a fully automatic segmentation pipelines [2] to annotate this data. The requirement of a very high level of accuracy and data rates approaching 1TB a day make this a challenging computer vision task. Deep Learning The first and most critical stage of the our segmentation pipeline (www.rhoana.org) is to identify membrane in the 2D images produced by the microscope. We use convolutional neural networks to classify images, trained on a few hand-annotated images. GPU computing greatly reduces the training and classification time for these computationally demanding networks. In this work, we used pylearn2 [5] to apply maxout networks [6] to two connectome data sets. -

Cryonics Magazine, September-October, 2012
September-October 2012 • Volume 33:5 Register for Alcor’s 40th Anniversary Conference Page 17 Symposium on Cryonics and Brain- Threatening Disorders Page 8 Book Review: Connectome: How the Brain’s Wiring Makes Us Who We Are Page 12 ISSN 1054-4305 $9.95 Improve Your Odds of a Good Cryopreservation You have your cryonics funding and contracts in place but have you considered other steps you can take to prevent problems down the road? _ Keep Alcor up-to-date about personal and medical changes. _ Update your Alcor paperwork to reflect your current wishes. _ Execute a cryonics-friendly Living Will and Durable Power of Attorney for Health Care. _ Wear your bracelet and talk to your friends and family about your desire to be cryopreserved. _ Ask your relatives to sign Affidavits stating that they will not interfere with your cryopreservation. _ Attend local cryonics meetings or start a local group yourself. _ Contribute to Alcor’s operations and research. Contact Alcor (1-877-462-5267) and let us know how we can assist you. Take a look at the Your source for news about: ALCOR BLOG • Cryonics technology • Cryopreservation cases • Television programs about cryonics http://www.alcor.org/blog/ • Speaking events and meetings • Employment opportunities Alcor Life Connect with Alcor members and supporters on our Extension official Facebook page: http://www.facebook.com/alcor.life.extension. Foundation foundation is on Become a fan and encourage interested friends, family members, and colleagues to support us too. CONTENTS 5 Quod incepimus lume 33:5