Reproducible Cells and Organoids Via Directed-Differentiation Encoding (RECODE) (Nsf21532) |
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Blueprint for Engineering Cell Fate: Current Technologies to Reprogram Cell Identity
Cell Research (2013) 23:33-48. © 2013 IBCB, SIBS, CAS All rights reserved 1001-0602/13 $ 32.00 npg REVIEW www.nature.com/cr A blueprint for engineering cell fate: current technologies to reprogram cell identity Samantha A Morris1, 2, 3, George Q Daley1, 2, 3 1Stem Cell Transplantation Program, Division of Pediatric Hematology and Oncology, Manton Center for Orphan Disease Re- search, Howard Hughes Medical Institute, Children’s Hospital Boston and Dana Farber Cancer Institute, Boston, MA, USA; 2De- partment of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA, USA; 3Harvard Stem Cell Institute, Cambridge, MA, USA Human diseases such as heart failure, diabetes, neurodegenerative disorders, and many others result from the deficiency or dysfunction of critical cell types. Strategies for therapeutic tissue repair or regeneration require the in vitro manufacture of clinically relevant quantities of defined cell types. In addition to transplantation therapy, the generation of otherwise inaccessible cells also permits disease modeling, toxicology testing and drug discovery in vitro. In this review, we discuss current strategies to manipulate the identity of abundant and accessible cells by dif- ferentiation from an induced pluripotent state or direct conversion between differentiated states. We contrast these approaches with recent advances employing partial reprogramming to facilitate lineage switching, and discuss the mechanisms underlying the engineering of cell fate. Finally, we address the current limitations of the field and how the resulting cell types can be assessed to ensure the production of medically relevant populations. Keywords: reprogramming; direct fate conversion; directed differentiation Cell Research (2013) 23:33-48. doi:10.1038/cr.2013.1; published online 1 January 2013 Introduction could only transition to progressively more differentiated states, with de-differentiation seen only in cases of tissue Cell differentiation has long been thought to be a uni- pathology (e.g., metaplasia or malignancy). -

Project Final Report
PROJECT FINAL REPORT Grant Agreement number: 229289 Project acronym: NANOII Project title: Nanoscopically-guided induction and expansion of regulatory hematopoietic cells to treat autoimmune and inflammatory processes Funding Scheme: Period covered: from month 1 to month 48 Name of the scientific representative of the project's co-ordinator, Title and Organisation: Prof. Dr. Joachim P. Spatz, Max-Planck-Institute, Stuttgart Tel: +49 711 689-3611 Fax: +49 711 689-3612 E-mail: [email protected] Project website address: http://www.mf.mpg.de/NanoII 1 4.1 Final publishable summary report Executive Summary NanoII Nanoscopically-guided induction and expansion of regulatory hematopoietic cells to treat autoimmune and inflammatory processes NanoII has been a colaborative project within the EU 7th framework program for research on Nanoscopically-guided induction and expansion of regulatory hematopoietic cells to treat autoimmune and inflammatory processes. This project has been developing novel approaches for directing the differentiaton, proliferation and tissue tropism of specific hematopoietic lineages, using micro-and nanofabricated cell chips. We are using advanced nanofabricated surfaces functionalized with specific biomolecules, and microfluidic cell chips to specify and expand regulatory immune cell for treating of important inflammatory and autoimmune disorders in an organ and antigen-specific manner. A new chip system creates ex vivo microenvironments mimicking in vivo cell-cell-interactions and molecular signals involved in differentiation and proliferation of hematopoietic cells. Key element of the project is the development of a novel high-throughput microscopy system for the identification of optimal conditions. The educated cells are employed in in-vivo experiments in new mouse models. -

NSF S2S Tip Sheet
Submission tips for system-to-system NSF proposals in RAMP 1. Proposals that do not include any committed effort/salary for the PI cannot be submitted system-to-system but must be submitted in either Fastlane or Research.gov. The Grants.gov application defaults adding the PI to the budget, but NSF does not allow anyone without salary to be included in the budget. 2. All solicitations should be reviewed to ensure Grants.gov submission is allowable before beginning an application. 3. The Biosketch and Other Support (Current and Pending) documents do not map to the SF424 for NSF applications, so it’s not necessary to upload them to the Personnel page in the Funding Proposal smartform. 4. NSF-Approved Formats must be used for the Biosketch and Current and Pending Support documents. If a different format is used, or the NSF template is altered in any way, the proposal will not upload successfully into Fastlane. The links for the approved templates can be found here: • https://www.nsf.gov/bfa/dias/policy/biosketch.jsp • https://www.nsf.gov/bfa/dias/policy/cps.jsp Last Revised 2/18/2021 5. RAMP does not flag an error if you are missing an additional performance site. You have to know if additional sites should be included (e.g. when you have subawards). 6. If a Performance Site is added, but the Organization Name is left blank, RAMP will return a non- descriptive error that will prevent the proposal from validating for submission. 7. Person month effort maps from the Proposal Budget to the SF424 as Calendar Months and may also calculate a number with several decimal places. -

Directed Differentiation of Human Embryonic Stem Cells
DirectedDirected differentiationdifferentiation ofof humanhuman embryonicembryonic stemstem cellscells TECAN Symposium 2008 Biologics - From Benchtop to Production Wednesday 17 September 2008 Andrew Elefanty Embryonic Stem Cell Differentiation Laboratory, MISCL ES cells as a system to dissect development hES ES cells derived from inner cell mass of preimplantation blastocyst Functionally similar, patient specific cells generated by reprogramming adult or fetal cells: • Somatic cell nuclear transfer (SCNT) - oocyte cytoplasm reprograms somatic cell • Induced pluripotential stem cell (iPS) – reprogramming is initiated by genes and/or growth factors introduced into adult cells ES cells can differentiate into all the cell types in the body blood cells differentiation liver heart muscle pancreas ES cell line nerve In vitro differentiation of embryonic stem cells provides an avenue to • study early development • generate tissue specific stem cells and mature end cells to study disease to screen drugs/therapeutic agents for cell therapies Directed differentiation of ES cells Recapitulate embryonic development in vitro to derive lineage precursors hES Mesendoderm Ectoderm Mesoderm Endoderm Factors that play key roles in embryogenesis also direct differentiation of ES cells hES BMP/Activin/Wnt/FGF Mesendoderm FGF/noggin BMP/Wnt Activin Ectoderm Mesoderm Endoderm Elements that facilitate directed differentiation of HESCs in vitro •large, uniform populations of undifferentiated HESCs •robust, reproducible differentiation protocol incorporating a -
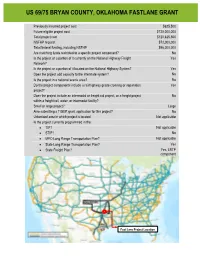
US 69/75 Upgrade to Limited Access Highway
US 69/75 BRYAN COUNTY, OKLAHOMA FASTLANE GRANT Previously incurred project cost $625,500 Future eligible project cost $120,000,000 Total project cost $120,625,500 NSFHP request $72,000,000 Total federal funding, including NSFHP $96,000,000 Are matching funds restricted to a specific project component? No Is the project or a portion of it currently on the National Highway Freight Yes Network? Is the project or a portion of it located on the National Highway System? Yes Does the project add capacity to the Interstate system? No Is the project in a national scenic area? No Do the project components include a rail/highway grade crossing or separation Yes project? Does the project include an intermodal or freight rail project, or a freight project No within a freight rail, water, or intermodal facility? Small or large project? Large Also submitting a TIGER grant application for this project? No Urbanized area in which project is located Not applicable Is the project currently programmed in the: TIP? Not applicable STIP? No MPO Long Range Transportation Plan? Not applicable State Long Range Transportation Plan? Yes State Freight Plan? Yes, LRTP component Fast Lane Project Location Fast Lane Project Location 1 US 69/75 BRYAN COUNTY, OKLAHOMA FASTLANE GRANT PROJECT NARRATIVE TABLE OF CONTENTS PROJECT DESCRIPTION ............................................................................................................................................. 1 PROJECT LOCATION ............................................................................................................................................... -

Health Services Research (Including System Process Change)
April 2013 Roadmap to the Research Hospital of the Future 1 Table of Contents INTRODUCTION ............................................................................................................. 3 KEY ASSUMPTIONS ...................................................................................................... 5 THEMES AND DEFINITIONS ......................................................................................... 6 TRENDS SUMMARY ...................................................................................................... 8 REGENERATIVE MEDICINE ........................................................................................ 11 EXPERIMENTAL THERAPEUTICS…………………..……………………………….…….19 MEDICAL TECHNOLOGY ............................................................................................ 29 INFORMATICS AND PATIENT INFORMATION COLLECTION & ASSESSMENT ...... 46 HEALTH SERVICES RESEARCH (INCLUDING SYSTEM PROCESS CHANGE) ....... 55 MECHANISMS OF DISEASE ....................................................................................... 73 RESEARCH AND EDUCATION ................................................................................... 89 APPENDIX .................................................................................................................... 94 GLOSSARY OF TERMS & ABBREVIATIONS ............................................................. 95 2 Introduction The UHN Research community undertook a major strategic planning exercise in 2002 in response to the release of the -

Nano-Biosensor for Monitoring the Neural Differentiation of Stem Cells
nanomaterials Review Nano-Biosensor for Monitoring the Neural Differentiation of Stem Cells Jin-Ho Lee 1,2,†, Taek Lee 1,2,† and Jeong-Woo Choi 1,2,* 1 Department of Chemical and Biomolecular Engineering, Sogang University, 35 Baekbeom-ro (Sinsu-dong), Mapo-gu, Seoul 121-742, Korea; [email protected] (J.-H.L.); [email protected] (T.L.) 2 Institute of Integrated Biotechnology, Sogang University, 35 Baekbeom-ro (Sinsu-dong), Mapo-gu, Seoul 121-742, Korea * Correspondence: [email protected]; Tel.: +82-2-718-1976; Fax: +82-2-3273-0331 † These authors contributed equally to this work. Academic Editors: Chen-Zhong Li and Ling-Jie Meng Received: 6 July 2016; Accepted: 17 November 2016; Published: 28 November 2016 Abstract: In tissue engineering and regenerative medicine, monitoring the status of stem cell differentiation is crucial to verify therapeutic efficacy and optimize treatment procedures. However, traditional methods, such as cell staining and sorting, are labor-intensive and may damage the cells. Therefore, the development of noninvasive methods to monitor the differentiation status in situ is highly desirable and can be of great benefit to stem cell-based therapies. Toward this end, nanotechnology has been applied to develop highly-sensitive biosensors to noninvasively monitor the neural differentiation of stem cells. Herein, this article reviews the development of noninvasive nano-biosensor systems to monitor the neural differentiation of stem cells, mainly focusing on optical (plasmonic) and eletrochemical methods. The findings in this review suggest that novel nano-biosensors capable of monitoring stem cell differentiation are a promising type of technology that can accelerate the development of stem cell therapies, including regenerative medicine. -

**Streaming-Live!* - 2021 Fastlane 21Th March 2021
**streaming-live!* - 2021 Fastlane 21th March 2021 WWE Fastlane 2021 Starting XI Live Video result for WWE Fastlane 2021 Live120 WWE Fastlane 2021 Live Stream HD VCU vs Video result for WWE Fastlane 2021 Live4231 WWE Fastlane 2021 PreMatch Build Up Ft James Video result for WWE Fastlane 2021 Live WATCH ONLINE WWE Fastlane 2021 Live Online WWE Fastlane 2021 Live WWE WWE 2020 Live Streams WWE Fastlane 2021 Live op tv WWE Fastlane 2021 Live Reddit WWE Fastlane 2021 Live 2020WWE2020 Strean WWE Fastlane 2021Live 25 November 2020 Broadcast Today USTV Live op tv WWE Fastlane 2021 Free On Tv Strean WWE Fastlane 2021Live score AEK Athens vs VCU Live WWE Fastlane 2021 Live Update Score WWE Fastlane 2021 Live AEK Athens vs WWE Fastlane 2021 Live WWE Fastlane 2021 Live WWE Fastlane 2021 Live on radio 2020 WWE Fastlane 2021 Live Start Time Today WWE Fastlane 2021 LIVE Latest team news lineups standardcouk › Sport › WWE 12 mins ago — WWE Fastlane 2021 LIVE The Gunners return to college WWE action this evening looking to bounce back from last weekends defeat A total of seven matches have been announced for the show. This includes four championship matches, two singles matches, and an intriguing intergender match.This event features a number of matches including the Intercontinental Championship between Big E and Apollo Crews, as well as the Women’s Tag Team Championship pitting Sasha Banks and Bianca Belair against defending champions Shayna Baszler and Nia Jax. 2021 Fastlane Start times Fastlane emanates from WWE’s ThunderDome, held in Florida’s Tropicana Field stadium. -

Biomimetic Coatings Obtained by Combinatorial Laser Technologies
coatings Review Biomimetic Coatings Obtained by Combinatorial Laser Technologies Emanuel Axente 1 , Livia Elena Sima 2 and Felix Sima 1,* 1 Center for Advanced Laser Technologies (CETAL), National Institute for Laser, Plasma and Radiation Physics (INFLPR), 409 Atomistilor, Magurele 077125, Romania; emanuel.axente@inflpr.ro 2 Department of Molecular Cell Biology, Institute of Biochemistry of the Romanian Academy, 296 Splaiul Independentei, Bucharest 060031, Romania; [email protected] * Correspondence: felix.sima@inflpr.ro; Tel.: +4-021-457-4243 Received: 13 April 2020; Accepted: 7 May 2020; Published: 9 May 2020 Abstract: The modification of implant devices with biocompatible coatings has become necessary as a consequence of premature loosening of prosthesis. This is caused mainly by chronic inflammation or allergies that are triggered by implant wear, production of abrasion particles, and/or release of metallic ions from the implantable device surface. Specific to the implant tissue destination, it could require coatings with specific features in order to provide optimal osseointegration. Pulsed laser deposition (PLD) became a well-known physical vapor deposition technology that has been successfully applied to a large variety of biocompatible inorganic coatings for biomedical prosthetic applications. Matrix assisted pulsed laser evaporation (MAPLE) is a PLD-derived technology used for depositions of thin organic material coatings. In an attempt to surpass solvent related difficulties, when different solvents are used for blending various organic materials, combinatorial MAPLE was proposed to grow thin hybrid coatings, assembled in a gradient of composition. We review herein the evolution of the laser technological process and capabilities of growing thin bio-coatings with emphasis on blended or multilayered biomimetic combinations. -

Sex-Dependent VEGF Expression Underlies Variations in Human Pluripotent Stem Cell to Endothelial Progenitor Differentiation
www.nature.com/scientificreports OPEN Sex-dependent VEGF expression underlies variations in human pluripotent stem cell to endothelial progenitor diferentiation Lauren N. Randolph1,2, Xiaoping Bao4, Michael Oddo1 & Xiaojun Lance Lian1,2,3* Human pluripotent stem cells (hPSCs) ofer tremendous promise in tissue engineering and cell-based therapies because of their unique combination of two properties: pluripotency and a high proliferative capacity. To realize this potential, development of efcient hPSC diferentiation protocols is required. In this work, sex-based diferences are identifed in a GSK3 inhibitor based endothelial progenitor diferentiation protocol. While male hPSCs efciently diferentiate into CD34 + CD31+ endothelial progenitors upon GSK3 inhibition, female hPSCs showed limited diferentiation capacity using this protocol. Using VE-cadherin-GFP knockin reporter cells, female cells showed signifcantly increased diferentiation efciency when treated with VEGF during the second stage of endothelial progenitor diferentiation. Interestingly, male cells showed no signifcant change in diferentiation efciency with VEGF treatment, but did show augmented early activation of VE-cadherin expression. A sex-based diference in endogenous expression of VEGF was identifed that is likely the underlying cause of discrepancies in sex-dependent diferentiation efciency. These fndings highlight the importance of sex diferences in progenitor biology and the development of new stem cell diferentiation protocols. In the regenerative medicine feld, topics such as personalized medicine, immunoengineering, and stem cell ther- apies are frequently discussed; however, cell sex variations are rarely considered despite prevalent evidence of sex diferences in many diferent diseases, such as cardiovascular disease, autoimmune disease, Alzheimer disease, and diabetes1–6. Many of these conditions involve malfunction or attack of terminally diferentiated senescent cells making them excellent candidates for the development of stem cell-based therapies. -

Reproducible Cells and Organoids Via Directed-Differentiation Encoding (RECODE)
This document has been archived and replaced by NSF 21-532. Reproducible Cells and Organoids via Directed-Differentiation Encoding (RECODE) PROGRAM SOLICITATION NSF 20-541 National Science Foundation Directorate for Engineering Division of Chemical, Bioengineering, Environmental and Transport Systems Letter of Intent Due Date(s) (required) (due by 5 p.m. submitter's local time): March 02, 2020 Full Proposal Deadline(s) (due by 5 p.m. submitter's local time): May 14, 2020 IMPORTANT INFORMATION AND REVISION NOTES Any proposal submitted in response to this solicitation should be submitted in accordance with the revised NSF Proposal & Award Policies & Procedures Guide (PAPPG) (NSF 20-1), which is effective for proposals submitted, or due, on or after June 1, 2020. SUMMARY OF PROGRAM REQUIREMENTS General Information Program Title: Reproducible Cells and Organoids via Directed- Differentiation Encoding (RECODE) Synopsis of Program: The National Science Foundation (NSF) Division of Chemical, Bioengineering, Environmental and Transport Systems (CBET), seeks proposals that elucidate mechanisms of, and develop strategies to, direct the differentiation of undifferentiated cells into mature, functional cells or organoids. Projects responsive to this solicitation must aim to establish a robust and reproducible set of differentiation design rules, predictive models, real-time sensing, control, and quality assurance methods, and integrate them into a workable differentiation strategy. They must develop a fundamental understanding of how cells develop, including mechanisms, molecular machinery, dynamics, and cell-cell interactions, and use this understanding to manipulate cells purposefully. Investigators can choose any undifferentiated cell type, from any animal species, as a starting point and choose any appropriate functional product (cell, organoid, etc.) with real-world relevance. -

A04120071 Page 1 of 1
+PC * S NATIONU SCIENCE FOUNDATION OFFICEOFFICE OF OF INSPECTOR INVESTIGATIONS GENERAL z4 .q$$@!0 CLOSEOUT MEMORANDUM D~~~~ \o Case Number: A04120071 Page 1 of 1 In connection with a proactive review, we reviewed an awardee's ' General Ledger. This review revealed that the awardee had spent the last $32,000 of NSF funds on post-award expenditures. Further iilvestigation revealed that the PI had requested an extension of tiine to complete work beyond the award's expiration, but NSF denied this request. In addition, investigation revealed that despite NSF's denial of the second-no-cost extension, the PI engineered the draw down of the $32,000 by falsely characteriziilg the draw down as a reimbursement. However, the ~najorityof the funds, approxi~nately$27,000, were used to fund newly incurred, ongoing, post-award expenses. Thereafter, the awardee filed a false final Federal Cash Transaction Report certifying that all award funds had been spent. Then, the PI filed a Final Report that falsely represented that all work on the award was complete. When asked about this by OIG investigators, the PI said she didn't know the second no-cost extension had been denied by NSF. The weight of the evidence refuted this assertion. Without adinitting liability, the awardee settled a civil false claims case with the Department of Justice for $52,150 and agreed to enter into a self-governance programdesigned to ensure that it will operate with honestly and integrity and in compliance with applicable law. In a separate and unrelated ad~ni~listrativeaction, the PI was debarred by NSF for 5 years.