Exploring Mechanistic Toxicity of Mixtures Using PBPK Modeling and Computational Systems Biology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Biological Safety Standard Operating Procedure (SOP) SOP Title
IBSP005 Biosafety SOP template January 2021 Icahn School of Medicine at Mount Sinai Biological Safety Standard Operating Procedure (SOP) SOP Title SOP Number (version) IBC registration(s) Section 1. Laboratory-specific information Building/Room(s) Department/Institute SOP author LSO PI name PI signature Section 2. Biological hazard information Biological Agent1 RG2 BSL3 Potential Hazards Signs/Symptoms of Occupational Health ABSL4 Infection Requirements Section 3. Other hazards Sharps Hazards Other Biological Hazards Chemical Hazards Radiological Hazards Section 4. Personal Protective Equipment (PPE) Laboratory Coat Fluid-Resistant Gown Surgical Mask Gloves Shoe Covers N95 Respirator Eye Protection Fluid-Resistant Sleeves PAPR Other PPE 1 Specific strains of biological agents should be described in the corresponding IBC registration. 2 NIH Risk Groups: RG-1, RG-2, RG-3, or RG-4 3 Biological Safety Level: BSL-1, BSL-2, BSL-3, BSL-4 4 Animal Biological Safety Level: ABSL-1, ABSL-2, ABSL-3, ABSL-4 Page | 1 IBSP005 Biosafety SOP template January 2021 Section 5. Equipment and Engineering Controls Biological Safety Cabinet Centrifuge Aerosol-Generating Equipment Other equipment Section 6. Decontamination Disinfectant Contact Time Dilution Location Section 7. Waste Management/Disposal Steam Sterilizer Location: (Autoclave) Chemical Disinfectant(s) Location: Other Disinfectant(s) Location: Section 8. Transport Procedure(s) Page | 2 IBSP005 Biosafety SOP template January 2021 Section 9. Spill Response Procedure Section 10. Protocol Procedure Page | 3 IBSP005 Biosafety SOP template January 2021 Section 10. Protocol Procedure Continued (attach additional sheets if necessary) Page | 4 IBSP005 Biosafety SOP template January 2021 Section 11. Documentation of training and understanding. The Principal Investigator must ensure that all laboratory personnel receive training on the content of this SOP. -

Occupational Safety and Health Admin., Labor § 1910.145
Occupational Safety and Health Admin., Labor § 1910.145 of the workers or their families, shall (i) Fire protection equipment and appa- be provided in connection with all food ratus. [Reserved] handling facilities. There shall be no (ii) Danger. Safety cans or other port- direct opening from living or sleeping able containers of flammable liquids quarters into a kitchen or dining hall. having a flash point at or below 80° F, (3) No person with any communicable table containers of flammable liquids disease shall be employed or permitted (open cup tester), excluding shipping to work in the preparation, cooking, containers, shall be painted red with serving, or other handling of food, food- some additional clearly visible identi- stuffs, or materials used therein, in fication either in the form of a yellow any kitchen or dining room operated in band around the can or the name of the connection with a camp or regularly contents conspicuously stenciled or used by persons living in a camp. painted on the can in yellow. Red (j) Insect and rodent control. Effective lights shall be provided at barricades measures shall be taken to prevent in- and at temporary obstructions, as spec- festation by and harborage of animal ified in ANSI Safety Code for Building or insect vectors or pests. Construction, A10.2±1944, which is in- (k) First aid. (1) Adequate first aid fa- corporated by reference as specified in cilities approved by a health authority § 1910.6. Danger signs shall be painted shall be maintained and made available red. in every labor camp for the emergency (iii) Stop. -
Lab Standard Operating Procedures (Sops)
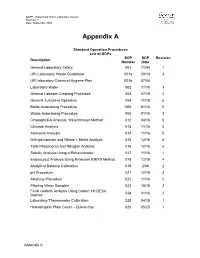
QAPP –Watershed Watch Laboratory Assays Revision: 7 Date: September 2020 Appendix A Standard Operation Procedures List of SOPs SOP SOP Revision Description Number Date General Laboratory Safety 001 11/04 1 URI Laboratory Waste Guidebook 001a 09/13 3 URI laboratory Chemical Hygiene Plan 001b 07/04 Laboratory Water 002 11/16 3 General Labware Cleaning Procedure 003 07/19 4 General Autoclave Operation 004 10/18 6 Bottle Autoclaving Procedure 005 01/16 5 Waste Autoclaving Procedure 006 01/16 3 Chlorophyll-A Analysis, Welschmeyer Method 012 04/18 6 Chloride Analysis 013 11/16 5 Ammonia Analysis 014 11/16 5 Orthophosphate and Nitrate + Nitrite Analysis 015 12/16 5 Total Phosphorus and Nitrogen Analysis 016 12/16 5 Salinity Analysis Using a Refractometer 017 11/16 1 Enterococci Analysis Using Enterolert IDEXX Method 018 12/19 4 Analytical Balance Calibration 019 2/06 2 pH Procedure 021 12/19 3 Alkalinity Procedure 022 11/16 2 Filtering Water Samples 023 10/18 2 Fecal coliform Analysis Using Colilert 18 IDEXX 024 01/16 2 Method Laboratory Thermometer Calibration 025 04/19 1 Heterotrophic Plate Count – Quanti-tray 026 05/20 1 Appendix A Standard Operating Procedure 001 Date: 11/04 General Laboratory Safety Revision: 1 Author: Linda Green University of Rhode Island Watershed Watch 1.0 PURPOSE AND DESCRIPTION LAB SAFETY IS EVERYBODY’S JOB! Please be sure to familiarize yourself with these general procedures, as well as the specific handling requirements included in the Standard Operating Procedure (SOP) for each analysis/process. Further general information regarding University of Rhode Island standards for health and safety are found in SOP 001a – University Safety and Waste Handling Document. -
Sampling Guide for First Responders to Drinking Water Contamination Threats and Incidents
Sampling Guide for First Responders to Drinking Water Contamination Threats and Incidents This First Responders Sampling Guide was funded under U.S. EPA Assistance Agreement No. X6-97109101-1. Sampling Guide for First Responders to Drinking Water Contamination Threats and Incidents New England Water Works Association 125 Hopping Brook Road Holliston, MA 01746-1471 November 2008 Acknowledgments This Sampling Guide for First Responders to Drinking Water Contamination Threats and Incidents was originally developed by the New England Water Works Association in collaboration with U.S. EPA. Please refer to pages 75 and 76 for a complete list of acknowledgments. The cover photo is reprinted with permission from the Worcester Telegram & Gazette. IMPORTANT NOTICE The methods and instructions in this guide reflect the U.S. Environmental Protection Agency (EPA) regulations and guidance, in addition to other reference documents. It is recognized that there can be significant differences from state to state regarding public notification, sampling procedures, laboratory safety, and handling, etc. Please check with your state drinking water representative with any questions before First Responders training and preparation is begun. Contents, Sections 1 through 6 1. Introduction 1.1 Background/Purpose ......................................1 1.2 How to Use This Guide ....................................2 2. Overview of Site Characterization .........................3 2.1 Investigating the Site .......................................3 2.2 Who Conducts Site Characterization and Sampling? .................................................5 3. Site Characterization Five-Step Process ...............6 3.1 The Five-Step Process .....................................6 Step 1: Customizing the Site Characterization Plan ........................8 Step 2: Approaching the Site and Doing a Field Safety Screening ........9 Step 3: Characterizing the Site ...................11 Step 4: Collecting Samples ..........................14 Step 5: Exiting the Site .................................16 4. -

Toxicogenomics Applications of New Functional Genomics Technologies in Toxicology
\-\w j Toxicogenomics Applications of new functional genomics technologies in toxicology Wilbert H.M. Heijne Proefschrift ter verkrijging vand egraa dva n doctor opgeza gva nd e rector magnificus vanWageninge n Universiteit, Prof.dr.ir. L. Speelman, in netopenbaa r te verdedigen op maandag6 decembe r200 4 des namiddagst e half twee ind eAul a - Table of contents Abstract Chapter I. page 1 General introduction [1] Chapter II page 21 Toxicogenomics of bromobenzene hepatotoxicity: a combined transcriptomics and proteomics approach[2] Chapter III page 48 Bromobenzene-induced hepatotoxicity atth etranscriptom e level PI Chapter IV page 67 Profiles of metabolites and gene expression in rats with chemically induced hepatic necrosis[4] Chapter V page 88 Liver gene expression profiles in relation to subacute toxicity in rats exposed to benzene[5] Chapter VI page 115 Toxicogenomics analysis of liver gene expression in relation to subacute toxicity in rats exposed totrichloroethylen e [6] Chapter VII page 135 Toxicogenomics analysis ofjoin t effects of benzene and trichloroethylene mixtures in rats m Chapter VII page 159 Discussion and conclusions References page 171 Appendices page 187 Samenvatting page 199 Dankwoord About the author Glossary Abbreviations List of genes Chapter I General introduction Parts of this introduction were publishedin : Molecular Biology in Medicinal Chemistry, Heijne etal., 2003 m NATO Advanced Research Workshop proceedings, Heijne eral., 2003 81 Chapter I 1. General introduction 1.1 Background /.1.1 Toxicologicalrisk -

Commodity Specific Food Safety Guidelines for the Fresh Tomato Supply Chain
COMMODITY SPECIFIC FOOD SAFETY GUIDELINES FOR THE FRESH TOMATO SUPPLY CHAIN Third Edition | September 2018 Commodity Specific Food Safety Guidelines for the Fresh Tomato Supply Chain Tomato Guidelines, 3rd Edition User’s Note These guidelines provide recommended food safety practices that are intended to minimize the microbiological hazards associated with fresh and fresh-cut tomato products. The intent of drafting this document is to provide currently available information on food safety and handling in a manner consistent with existing applicable regulations, standards and guidelines. The information provided herein is offered in good faith and believed to be reliable, but is made without warranty, express or implied, as to merchantability, fitness for a particular purpose, or any other matter. These recommended guidelines were not designed to apply to any specific operation. It is the responsibility of the user of this document to verify that these guidelines are appropriate for its operation. The publishing trade associations, their members and contributors do not assume any responsibility for compliance with applicable laws and regulations, and recommend that users consult with their own legal and technical advisers to be sure that their own procedures meet with applicable requirements. - 1 - Foreword The North American Tomato Trade Work Group (NATTWG) published in 2006 the first edition of Commodity Specific Food Safety Guidelines for the Fresh Tomato Supply Chain (“Guidelines”). Within two years of publication, several initiatives resulted in significant new learnings about potential risks and control measures at all points in the fresh tomato supply chain. Some of those initiatives include the FDA Tomato Safety Initiative, voluntary efforts by the Florida Tomato Exchange and the California Tomato Farmers to develop USDA-verified audit criteria and programs for tomato production and harvest practices in those states, and several retail and foodservice buyer initiatives to further define tomato safe growing and handling practices. -

Genomic Dose Response
Genomic Dose Response: The Picture NTP Genomic Dose Response Modeling Expert Panel Meeting October 23, 2017 Russell Thomas Director National Center for Computational Toxicology The views expressed in this presentation are those of the author and do not necessarily reflect the views or policies of the U.S. EPA Scott May Want To Rethink Asking Me To Be The “Big Picture Guy”… National Center for Computational Toxicology It is a Well Known Fact that Toxicology Continues to Have a Data Problem US National Research Council, 1984 Size of Estimate Mean Percent Category Category In the Select Universe Pesticides and Inert Ingredients of Pesticides 3,350 Formulations 10 24 2 26 38 Cosmetic Ingredients 3,410 2 14 10 18 56 Drugs and Excipients Used in Drug Formulations 1,815 18 18 3 36 25 Food Additives 8,627 • Major challenge is too many 5 14 1 34 46 Chemicals in Commerce: chemicals and not enough At Least 1 Million 12,860 Pounds/Year data 11 11 78 Chemicals in Commerce: • Total # chemicals = 65,725 Less than 1 Million 13,911 Pounds/Year • Chemicals with no toxicity 12 12 76 data of any kind = ~46,000 Chemicals in Commerce: Production Unknown or 21,752 Inaccessible 10 8 82 Complete Partial Minimal Some No Toxicity Health Health Toxicity Toxicity Information Hazard Hazard Information Information Available Assessment Assessment Available Available 2 Possible Possible (But Below Minimal) National Center for Computational Toxicology It is a Well Known Fact that Toxicology Continues to Have a Data Problem 70 60 50 40 30 20 Percent of Chemicals <1% 10 0 Acute Cancer Gentox Dev Tox Repro Tox EDSP Tier 1 Modified from Judson et al., EHP 2009 3 National Center for Computational Toxicology For Those With Data, Have We Been Truly Predictive or Just Protective? …data compiled from 150 compounds with 221 human toxicity events reported. -

Finding Hazards
FINDING HAZARDS OSHA 11 Finding Hazards 1 Osha 11 Finding Hazards 2 FINDING HAZARDS Learning Objectives By the end of this lesson, students will be able to: • Define the term “job hazard” • Identify a variety of health and safety hazards found at typical worksites where young people are employed. • Locate various types of hazards in an actual workplace. Time Needed: 45 Minutes Materials Needed • Flipchart Paper • Markers (5 colors per student group) • PowerPoint Slides: #1: Job Hazards #2: Sample Hazard Map #3: Finding Hazards: Key Points • Appendix A handouts (Optional) Preparing To Teach This Lesson Before you present this lesson: 1. Obtain a flipchart and markers or use a chalkboard and chalk. 2. Locate slides #1-3 on your CD and review them. If necessary, copy onto transparencies. 3. For the Hazard Mapping activity, you will need flipchart paper and a set of five colored markers (black, red, green, blue, orange) for each small group. Detailed Instructor’s Notes A. Introduction: What is a job hazard? (15 minutes) 1. Remind the class that a job hazard is anything at work that can hurt you, either physically or mentally. Explain that some job hazards are very obvious, but others are not. In order to be better prepared to be safe on the job, it is necessary to be able to identify different types of hazards. Tell the class that hazards can be divided into four categories. Write the categories across the top of a piece of flipchart paper and show PowerPoint Slide #1, Job Hazards. • Safety hazards can cause immediate accidents and injuries. -

Next-Generation Sequencing Data for Use in Risk Assessment Bruce Alexander Merrick
Available online at www.sciencedirect.com Current Opinion in ScienceDirect Toxicology Next-generation sequencing data for use in risk assessment Bruce Alexander Merrick Abstract fluorescently labeled nucleotides and capillary electro- Next-generation sequencing (NGS) represents several phoresis that gave rise to automated sequencing in- powerful platforms that have revolutionized RNA and DNA struments such as the Applied Biosystems, Inc. model analysis. The parallel sequencing of millions of DNA molecules 370A sequencers and others, on the basis of Sanger can provide mechanistic insights into toxicology and provide chemistries [2]. Read length per each sample was at 500e new avenues for biomarker discovery with growing relevance 800 nucleotides, and sample throughput was limited. for risk assessment. The evolution of NGS technologies has improved over the last decade with increased sensitivity and Sanger-based DNA sequencing instruments are accuracy to foster new biomarker assays from tissue, blood, considered first-generation platforms. Instruments that and other biofluids. NGS technologies can identify transcrip- perform multiple sequencing reactions simultaneously tional changes and genomic targets with base pair precision in in a ‘massively parallel’ fashion have been dubbed, response to chemical exposure. Furthermore, there are ‘NextGeneration’ or NextGen Sequencing [3]. How several exciting movements within the toxicology community NGS came about compared with other genomic plat- that incorporate NGS platforms into new strategies for more forms is interlinked with microarray technology rapid toxicological characterizations. These include the Tox21 (Figure 1). Both platforms can provide whole genomic in vitro high-throughput transcriptomic screening program, approaches to research problems. Microarrays are a development of organotypic spheroids, alternative animal fluorescent probe hybridization-based technology with models, mining archival tissues, liquid biopsy, and epige- origins in the mid-1990s that are now a mature genomic nomics. -

Working with Biological Agents (PDF, 564KB)
Working with biological agents Guidance notes These Notes provide guidance on aspects of work with biological agents. Several contain MRC Codes of Practice (CoP). Those containing a CoP are marked with * in the index below. Where a CoP is specified, establishments are expected to achieve and maintain those standards, which are set as the minimum acceptable for carrying out the related work. The associated guidance will aid the achievement of MRC standards and ensure the work meets all related legal requirements. In all MRC CoPs, 'must' indicates a mandatory requirement whereas 'should' reflects a strong recommendation. Statements using should relate to the required standard and any modification must be justified through the process of a suitable and sufficient risk assessment. Where appropriate the content of these notes can be adopted as local codes of practice and referred to by risk assessments. Establishments are encouraged however to adapt the content for local needs, with the strict provision that local standards are at least as high as those stipulated in this guidance. Part A The management of work with biological material Guidance note 1 a) The appointment and duties of a Biological Safety Officer: b) The formation and remit of Biological Safety Committees Guidance note 2 Laboratory management Guidance note 3 Risk assessments and related procedures - overview Guidance note 4 Use of blood from volunteer staff, including use of ‘own’ cells* Part B Working with biological hazards Guidance note 5 Working with human pathogens* Guidance note 6 Working with transmissible spongiform encephalopathies* Guidance note 7 Working with genetically modified organisms Guidance Note 8 Working with cell cultures Guidance Note 9 Working with human material: 1. -

Personal Protective Equipment
LABORATORY BIOSAFETY MANUAL FOURTH EDITION AND ASSOCIATED MONOGRAPHS PERSONAL PROTECTIVE EQUIPMENT LABORATORY BIOSAFETY MANUAL FOURTH EDITION AND ASSOCIATED MONOGRAPHS PERSONAL PROTECTIVE EQUIPMENT Personal protective equipment (Laboratory biosafety manual, fourth edition and associated monographs) ISBN 978-92-4-001141-0 (electronic version) ISBN 978-92-4-001142-7 (print version) © World Health Organization 2020 Some rights reserved. This work is available under the Creative Commons Attribution- NonCommercial-ShareAlike 3.0 IGO licence (CC BY-NC-SA 3.0 IGO; https://creativecommons.org/ licenses/by-nc-sa/3.0/igo). Under the terms of this licence, you may copy, redistribute and adapt the work for non-commercial purposes, provided the work is appropriately cited, as indicated below. In any use of this work, there should be no suggestion that WHO endorses any specific organization, products or services. The use of the WHO logo is not permitted. If you adapt the work, then you must license your work under the same or equivalent Creative Commons licence. If you create a translation of this work, you should add the following disclaimer along with the suggested citation: “This translation was not created by the World Health Organization (WHO). WHO is not responsible for the content or accuracy of this translation. The original English edition shall be the binding and authentic edition”. Any mediation relating to disputes arising under the licence shall be conducted in accordance with the mediation rules of the World Intellectual Property Organization (http://www.wipo.int/amc/en/ mediation/rules/). Suggested citation. Personal protective equipment. Geneva: World Health Organization; 2020 (Laboratory biosafety manual, fourth edition and associated monographs). -

Complete Manual Cdc-Pdf
Framework for Assessing Health Impacts of Multiple Chemicals and Other Stressors (Update) U.S. Department of Health and Human Services Public Health Service Agency for Toxic Substances and Disease Registry Division of Toxicology February 2018 ii PREFACE The mission of the Agency for Toxic Substances and Disease Registry (ATSDR) is to serve the public by using the best science, taking responsive public health actions, and providing trusted health information to prevent harmful exposures and disease related to toxic substances. The U.S. Comprehensive Environmental Response, Compensation, and Liability Act (CERCLA, also known as the Superfund act) mandates that ATSDR determine whether people living near or at a hazardous waste site are being exposed, have been exposed, or will be exposed to toxic substances, whether that exposure is harmful, and what can be done to stop or reduce harmful exposures. CERCLA requires that ATSDR consider the following factors when evaluating the possible public health impacts of communities near Superfund sites: (1) the nature and extent of contamination at a site; (2) the demographics of the site population; (3) exposure pathways that may exist at a site (the extent to which people contact site contaminants); and (4) health effects and disease-related data. In addition, ATSDR is also authorized to conduct public health assessments at storage, treatment, and disposal facilities for hazardous wastes when requested by EPA, under the 1984 amendments to the Resource Conservation and Recovery Act of 1976 (RCRA). In addition, ATSDR conducts public health assessments for toxic substances, when petitioned by concerned community members, physicians, state or federal agencies, or tribal governments.