A Synthetic Biology Application in Metabolic Engineering by Nikolaos
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Metabolic Engineering of Microorganisms for the Overproduction of Fatty Acids Ting Wei Tee Iowa State University
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2013 Metabolic engineering of microorganisms for the overproduction of fatty acids Ting Wei Tee Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Chemical Engineering Commons Recommended Citation Tee, Ting Wei, "Metabolic engineering of microorganisms for the overproduction of fatty acids" (2013). Graduate Theses and Dissertations. 13516. https://lib.dr.iastate.edu/etd/13516 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Metabolic engineering of microorganisms for the overproduction of fatty acids by Ting Wei Tee A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of Doctor of Philosophy Major: Chemical Engineering Program of Study Committee: Jacqueline V. Shanks, Major Professor Laura R. Jarboe R. Dennis Vigil David J. Oliver Marna D. Nelson Iowa State University Ames, Iowa 2013 Copyright © Ting Wei Tee, 2013. All rights reserved. ii TABLE OF CONTENTS Page ACKNOWLEDGEMENTS ................................................................................................ v ABSTRACT ....................................................................................................................... -

Metabolic Engineering and Synthetic Biology-CHEN 4803 Chemical and Biological Engineering Fall 2016 Syllabus
Metabolic Engineering and Synthetic Biology-CHEN 4803 Chemical and Biological Engineering Fall 2016 Syllabus Importance of Course: Metabolic Engineering describes the field of study concerned with applying genetic engineering tools to alter flux through native or newly introduced metabolic pathways in biological systems. Synthetic Biology includes similar objectives but is more broadly focused on applications enabled more generally by advances in DNA synthesis and sequencing technologies. Together these fields are of central importance to efforts to produce chemicals, fuels, and materials via bioprocessing as well as a much broader range of emerging commercial applications; such as biosensing, consumer biotechnology, the microbiome, novel pharmaceuticals, etc. Objectives: With your help, make this the class you find most interesting and useful for your future career. See Course Learning Goals at the end of this syllabus for specifics. Expectations: I expect you to attend all classes and be on time. I expect you to be responsible for your learning in this class. However, I also expect you to come to me with difficult to answer questions. The major responsibility for learning must be by doing the assigned readings and being actively engaged in class discussions, workshops, and interactive presentations. Course Information Instructor: Dr. Anushree Chatterjee, Assistant Professor, Chemical and Biological Engineering, [email protected] Class Meetings: MWF, 11:30 AM-12:20 PM, BIOT A104 Textbook: None specific, though we will be following some parts of Metabolic Engineering: Principles and Methodologies by Gregory N. Stephanopoulus, Aristos A. Aristidou, and Jens Nielsen. Readings: Will be posted on D2L Course webpage or provided as handouts in class. -

Production of Succinic Acid by E.Coli from Mixtures of Glucose
2005:230 CIV MASTER’S THESIS Production of Succinic Acid by E. coli from Mixtures of Glucose and Fructose ANDREAS LENNARTSSON MASTER OF SCIENCE PROGRAMME Chemical Engineering Luleå University of Technology Department of Chemical Engineering and Geosciences Division of Biochemical and Chemical Engineering 2005:230 CIV • ISSN: 1402 - 1617 • ISRN: LTU - EX - - 05/230 - - SE Abstract Succinic acid, derived from fermentation of renewable feedstocks, has the possibility of replacing petrochemicals as a building block chemical. Another interesting advantage with biobased succinic acid is that the production does not contribute to the accumulation of CO2 to the environment. The produced succinic acid can therefore be considered as a “green” chemical. The bacterium used in this project is a strain of Escherichia coli called AFP184 that has been metabolically engineered to produce succinic acid in large quantities from glucose during anaerobic conditions. The objective with this thesis work was to evaluate whether AFP184 can utilise fructose, both alone and in mixtures with glucose, as a carbon source for the production of succinic acid. Hydrolysis of sucrose yields a mixture of fructose and glucose in equal ratio. Sucrose is a common sugar and the hydrolysate is therefore an interesting feedstock for the production of succinic acid. Fermentations with an initial sugar concentration of 100 g/L were conducted. The sugar ratios used were 100 % fructose, 100 % glucose and a mixture with 50 % fructose and glucose, respectively. The fermentation media used was a lean, low- cost media based on corn steep liquor and a minimal addition of inorganic salts. Fermentations were performed with a 12 L bioreactor and the acid and sugar concentrations were analysed with an HPLC system. -

Metabolic Engineering for Unusual Lipid Production in Yarrowia Lipolytica
microorganisms Review Metabolic Engineering for Unusual Lipid Production in Yarrowia lipolytica Young-Kyoung Park * and Jean-Marc Nicaud Micalis Institute, AgroParisTech, INRAE, Université Paris-Saclay, 78352 Jouy-en-Josas, France; [email protected] * Correspondence: [email protected]; Tel.: +33-(0)1-74-07-16-92 Received: 14 November 2020; Accepted: 2 December 2020; Published: 6 December 2020 Abstract: Using microorganisms as lipid-production factories holds promise as an alternative method for generating petroleum-based chemicals. The non-conventional yeast Yarrowia lipolytica is an excellent microbial chassis; for example, it can accumulate high levels of lipids and use a broad range of substrates. Furthermore, it is a species for which an array of efficient genetic engineering tools is available. To date, extensive work has been done to metabolically engineer Y. lipolytica to produce usual and unusual lipids. Unusual lipids are scarce in nature but have several useful applications. As a result, they are increasingly becoming the targets of metabolic engineering. Unusual lipids have distinct structures; they can be generated by engineering endogenous lipid synthesis or by introducing heterologous enzymes to alter the functional groups of fatty acids. In this review, we describe current metabolic engineering strategies for improving lipid production and highlight recent researches on unusual lipid production in Y. lipolytica. Keywords: Yarrowia lipolytica; oleochemicals; lipids; unusual lipids; metabolic engineering 1. Introduction Microbial lipids are promising alternative fuel sources given growing concerns about climate change and environmental pollution [1–4]. They offer multiple advantages over plant oils and animal fats. For example, the production of microbial lipids does not result in resource competition with food production systems; is largely independent of environmental conditions; can be based on diverse substrates; and allows product composition to be customized based on the desired application [3,4]. -

Metabolic Engineering Biological Art of Producing Useful Chemicals
GENERAL ARTICLE Metabolic Engineering Biological Art of Producing Useful Chemicals Ram Kulkarni Metabolic engineering is a process for modulating the me- tabolism of the organisms so as to produce the required amounts of the desired metabolite through genetic manipula- tions. Considering its advantages over the other chemical synthesis routes, this area of biotechnology is likely to revolu- tionize the way in which commodity chemicals are produced. RamRam KulkarniKulkarni isis anan AssistantAittPf Professor at t Introduction Symbiosis International University,Pune.His Living organisms have numerous biochemical reactions operat- research interests are in ing in them. These reactions allow the organisms to survive by metabolic engineering of processes such as generation of energy, production of fundamen- lactic acid bacteria for tal building blocks required for structural organization and syn- increasing their nutraceutical properties. thesis of biomolecules having specialized functions. Some of the chemicals generated during this process (called metabolism), are useful to mankind for various applications. These so-called value- added chemicals include various bioactive secondary metabolites such as an anti-malarial drug (artemesinin), chemicals required as the raw material for the synthesis of other molecules such as lactic acid, chemicals imparting flavor to food material such as terpe- 1 Racemic mixtures contain nes, biofuels and associated chemicals such as ethanol and bu- equal concentrations of mirror tanol, etc. The traditional way of utilization of such chemicals is image isomers (enantiomers) of that optically active chemical. to cultivate the host organisms producing these chemicals fol- lowed by harvesting the desired biochemical. However, in many cases, the quantities of the useful chemicals in the cells are very low, thus demanding cultivation of the organisms on a very large scale. -

Complete Thesis
University of Groningen Synthetic biology tools for metabolic engineering of the filamentous fungus Penicillium chrysogenum Polli, Fabiola IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please check the document version below. Document Version Publisher's PDF, also known as Version of record Publication date: 2017 Link to publication in University of Groningen/UMCG research database Citation for published version (APA): Polli, F. (2017). Synthetic biology tools for metabolic engineering of the filamentous fungus Penicillium chrysogenum. University of Groningen. Copyright Other than for strictly personal use, it is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), unless the work is under an open content license (like Creative Commons). The publication may also be distributed here under the terms of Article 25fa of the Dutch Copyright Act, indicated by the “Taverne” license. More information can be found on the University of Groningen website: https://www.rug.nl/library/open-access/self-archiving-pure/taverne- amendment. Take-down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Downloaded from the University of Groningen/UMCG research database (Pure): http://www.rug.nl/research/portal. For technical reasons the number of authors shown on this cover page is limited to 10 maximum. Download date: 06-10-2021 Synthetic biology tools for metabolic engineering of the filamentous fungus Penicillium chrysogenum PhD thesis to obtain the degree of PhD at the University of Groningen on the authority of the Rector Magnificus Prof. -

Metabolic Engineering Strategies for Butanol Production in Escherichia Coli
Received: 13 May 2019 | Revised: 3 April 2020 | Accepted: 4 May 2020 DOI: 10.1002/bit.27377 REVIEW Metabolic engineering strategies for butanol production in Escherichia coli Sofia Ferreira1,2 | Rui Pereira3,4 | S. A. Wahl5 | Isabel Rocha1,2 1CEB‐Centre of Biological Engineering, University of Minho, Campus de Gualtar, Braga, Portugal 2Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa (ITQB‐NOVA), Oeiras, Portugal 3SilicoLife Lda, Braga, Portugal 4Department of Biology and Biological Engineering, Chalmers University of Technology, Gothenburg, Sweden 5Department of Biotechnology, Delft University of Technology, Delft, The Netherlands Correspondence Isabel Rocha, CEB‐Centre of Biological Abstract Engineering, University of Minho, Campus de Gualtar, 4710‐057 Braga, Portugal. The global market of butanol is increasing due to its growing applications as solvent, Email: [email protected] flavoring agent, and chemical precursor of several other compounds. Recently, the Present address superior properties of n‐butanol as a biofuel over ethanol have stimulated even Sofia Ferreira, Instituto de Tecnologia Química more interest. (Bio)butanol is natively produced together with ethanol and acetone e Biológica António Xavier, Universidade Nova de Lisboa (ITQB‐NOVA), Oeiras, Portugal. by Clostridium species through acetone‐butanol‐ethanol fermentation, at non- competitive, low titers compared to petrochemical production. Different butanol Funding information Ministério da Ciência, Tecnologia e Ensino production pathways have been expressed in Escherichia coli, a more accessible host Superior, Grant/Award Number: ERA‐IB‐2/ compared to Clostridium species, to improve butanol titers and rates. The biopro- 0002/2014; Fundação para a Ciência e a Tecnologia, Grant/Award Numbers: ERA‐IB‐2/ duction of butanol is here reviewed from a historical and theoretical perspective. -

Multidimensional Engineering for the Production of Fatty Acid Derivatives in Saccharomyces Cerevisiae
THESIS FOR DEGREE OF DOCTOR OF PHILOSOPHY Multidimensional engineering for the production of fatty acid derivatives in Saccharomyces cerevisiae YATING HU Department of Biology and Biological Engineering CHALMERS UNIVERSITY OF TECHNOLOGY Gothenburg, Sweden 2019 Multidimensional engineering for the production of fatty acid derivatives in Saccharomyces cerevisiae YATING HU ISBN 978-91-7905-174-7 © YATING HU, 2019. Doktorsavhandlingar vid Chalmers tekniska högskola Ny serie nr 4641 ISSN 0346-718X Department of Biology and Biological Engineering Chalmers University of Technology SE-412 96 Gothenburg Sweden Telephone + 46 (0)31-772 1000 Cover: Multidimensional engineering strategies enable yeast as the cell factory for the production of diverse products. Printed by Chalmers Reproservice Gothenburg, Sweden 2019 Multidimensional engineering for the production of fatty acid derivatives in Saccharomyces cerevisiae YATING HU Department of Biology and Biological Engineering Chalmers University of Technology Abstract Saccharomyces cerevisiae, also known as budding yeast, has been important for human society since ancient time due to its use during bread making and beer brewing, but it has also made important contribution to scientific studies as model eukaryote. The ease of genetic modification and the robustness and tolerance towards harsh conditions have established yeast as one of the most popular chassis in industrial-scale production of various compounds. The synthesis of oleochemicals derived from fatty acids (FAs), such as fatty alcohols and alka(e)nes, has been extensively studied in S. cerevisiae, which is due to their key roles as substitutes for fossil fuels as well as their wide applications in other manufacturing processes. Aiming to meet the commercial requirements, efforts in different engineering approaches were made to optimize the TRY (titer, rate and yield) metrics in yeast. -
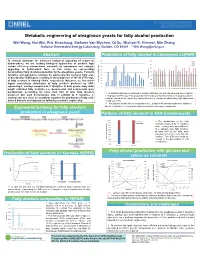
Metabolic Engineering of Oleaginous Yeasts for Fatty Alcohol Production Wei Wang, Hui Wei, Eric Knoshaug, Stefanie Van Wychen, Qi Xu, Michael E
Metabolic engineering of oleaginous yeasts for fatty alcohol production Wei Wang, Hui Wei, Eric Knoshaug, Stefanie Van Wychen, Qi Xu, Michael E. Himmel, Min Zhang National Renewable Energy Laboratory, Golden, CO 80401 * [email protected] Abstract Production of fatty alcohol in Lipomyces Ls[FAR] 900 70.0 To develop pathways for advanced biological upgrading of sugars to C16 C18 800 hydrocarbons, we are seeking biological approaches to produce high 60.0 C18:1 C20 carbon efficiency intermediates amenable to separations and catalytic 700 50.0 upgrading to hydrocarbon fuels. In this study, we successfully 600 demonstrated fatty alcohols production by the oleaginous yeasts, Yarrowia 500 40.0 400 lipolytica and Lipomyces starkeyi, by expressing the bacterial fatty acyl- 30.0 CoA reductase (FAR) gene, resulting in the production of 167 and 770 mg/L 300 Fatty alcohol(%) 20.0 200 of fatty alcohols in shaking flasks, respectively. Moreover, we find much (mg/L) production alcohol Fatty higher extracellular distribution of fatty alcohols produced by FAR- 100 10.0 expressing L. starkeyi compared to Y. lipolytica. In both yeasts, long chain 0 0.0 C t1 t2 t3 t4 t5 t6 t7 t8 t9 t10 t11 t12 t13 t14 t15 t16 t17 t18 t19 t20 length saturated fatty alcohols, i.e., hexadecanol and octadecanol were t1 t2 t3 t4 t5 t6 t7 t8 t9 t10 t11 t12 t13 t14 t15 t16 t17 t18 t19 t20 predominant, accounting for more than 85% of total fatty alcohols 70 transformants were confirmed to produce fatty alcohols with varied levels, among which produced. Our work demonstrates that, in addition to Y. -

Sugar Pathways in Fungi and Metabolic Engineering As a Tool for Biochemical Production
Outi Koivistoinen, M.Sc. VTT Technical Research Centre of Finland [email protected] Sugar pathways in fungi and metabolic engineering as a tool for biochemical production Currently the production of a vast majority of fuels and chemicals is still based on crude oil. Depletion of fossil fuels and the concern about carbon dioxide emissions are issues needing serious attention in the near future. Therefore alternative, more sustainable ways for efficient production of fuels and chemicals are needed. An ideal way for production would be a Biorefinery based on waste materials. Many of the desired chemicals to be produced by microbes used as production hosts in Biorefineries are not needed by the microbe; therefore they are not produced at all or the production yields are not sufficient enough for industrial purposes. To increase production rates or to produce new compounds metabolic engineering of the host organism is needed. New pathways providing the enzymes to convert the raw material to the desired chemical can be introduced to host cells, or pathways already existing can be boosted up, or attenuated to redirect the carbon provided. In order to be able to engineer the cells, profound understanding of the host metabolism is needed. Genes expressing the enzymes of various metabolic pathways need to be characterized and regulators behind the repression and activation mechanisms need to be unravelled. Host selection is an important consideration in developing production organisms for Biorefineries, and fungi are often preferred as they possess many desired properties. Yeast Saccharomyces cerevisiae is well known as an ethanol producer and methods to engineer it are well in place. -

Metabolic Engineering of Escherichia Coli for Itaconate Production
M e t a b o l i c Metabolic engineering of Escherichia coli e n g i n for itaconate production e e r i n g o f E . Kiira S. Vuoristo c o l i f o r i t a c o n a t e p r o d u c t i o n K i i r a S . V u o r i s t o Metabolic engineering of Escherichia coli for itaconate production Kiira S. Vuoristo Thesis committee Promotors Prof. Dr Gerrit Eggink Professor of Industrial Biotechnology Wageningen University Prof. Dr Johan P.M. Sanders Emeritus Professor of Valorisation of Plant Production Chains Wageningen University Co-promotor Dr Ruud A. Weusthuis Associate professor, Bioprocess Engineering Group Wageningen University Other members Prof. Dr Marjan De Mey, University of Gent, Belgium Prof. Dr Henk Noorman, DSM, Delft Prof. Dr Merja Penttilä, VTT Technical Research Centre of Finland, Oulu, Finland Prof. Dr Fons Stams, Wageningen University This research was conducted under the auspices of the Graduate School VLAG (Advanced studies in Food Technology, Agrobiotechnology, Nutrition and Health Sciences). Metabolic engineering of Escherichia coli for itaconate production Kiira S. Vuoristo Thesis submitted in fulfilment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. Dr A.P.J. Mol in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Friday 12 February 2016 at 4 p.m. in the Aula. Kiira S. Vuoristo Metabolic engineering of Escherichia coli for itaconate production 162 pages PhD thesis, Wageningen University, Wageningen, NL (2016) -

1 CM4125 Bioprocess Engineering Lab: Week 4
CM4125 Bioprocess Engineering Lab: Week 4: Introduction to Metabolic Engineering and Metabolic Flux Analysis Instructors: David R. Shonnard1, Susan T. Bagley2 Laboratory Teaching Assistant: Abraham Martin-Garcia 1 Chemical Engineering, 2 Biological Sciences Michigan Technological University, Houghton, MI 49931 January 31, 2006 102 Chemical Sciences and Engineering Building 1 Presentation Overview n Introduction to Metabolic Engineering n An Example of Metabolic Engineering: Ethanol Production from Lignocellulosic Biomass Using a Genetically-Engineered E. coli. n Metabolic Flux Analysis (MFA) n Defining the Reaction Stoichiometry in MFA n Defining the Reaction Rate Equations in MFA n Summary 2 What is Metabolic Engineering? “the directed improvement of product formation or cellular properties through the modification of specific biochemical reactions or the introduction of new ones with the use of recombinant DNA technology” Stephanopoulos, et al., 1998, Metabolic Engineering: Principles and Methodologies, Academic Press The targeting of specific biochemical reactions within the cell for modification is the essential feature of metabolic engineering. To achieve this targeting, we use Metabolic Flux Analysis (MFA). 3 Old Practice of Metabolic Engineering The concept of manipulating cellular metabolism for the purpose of increasing production of a desired product is an old one. Microoganisms have been developed with enhanced metabolic features. • DNA mutations using chemical treatment • Selection techniques to identify strains • Notable