NIH Public Access Author Manuscript Curr Pharm Biotechnol
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

FELLOW, ONCOLOGY RESEARCH – Southern Research, Birmingham, AL SOUTHERN RESEARCH, a Nonprofit, Scientific Research Organization
FELLOW, ONCOLOGY RESEARCH – Southern Research, Birmingham, AL SOUTHERN RESEARCH, a nonprofit, scientific research organization, is seeking an outstanding scientist for the position of Fellow in the field of Oncology The position is equivalent to the rank of Assistant, Associate or Full Professor in an academic institution (depending on the incumbent’s experience) and offers a unique opportunity to join the growing research team at Southern Research. Southern Research promotes a collaborative research environment to facilitate the multidisciplinary nature of drug discovery, accomplished internally through integration of basic biology, translational research, chemistry, and high-throughput screening, and also through its regional, national and international academic and industrial partnerships. The Oncology Department is focused on combining novel basic research and discovery to identify next generation cancer therapeutics. In contrast to traditional academic environments, Southern Research’s Oncology Department offers the opportunity for the individual to focus exclusively on engaging in research and professional development activities for advancement and development of new research ideas, support for the research team, and overall ability to make innovative advances in scholarly work portfolios. The Fellow of Oncology requires distinguished scholarship, including 10+ years of research experience in the field of oncology from any one of a number of concentrations, including cancer immunotherapy, cancer stem cells, cancer genomics, cancer pharmacogenomics, cancer microbiome, cancer pharmacology, genomic instability, or closely related area, and an ongoing extramurally-funded research program related to drug discovery. Southern Research desires a collegial, collaborative and outstanding scientist to cultivate mentorship and lead research within the Oncology Department. To be considered, applicants must: 1) possess a Ph.D., M.D. -

Pathology in Focus
PATHOLOGY IN FOCUS Vol 5 Issue 3 August 2011 Message from the Chair The start of the many faculty candidates will be several Department of Pathology Inside this issue: new school year visiting UAB and I hope that you will investigators are tentatively Faculty Profile: Dr. Hébert- 2-3 once again brings be able to attend as many candi- scheduled to move this fall into Magee many new faces date seminars as possible. renovated laboratory space in the to the Department Wallace Tumor Institute. These APC Teaching Awards 3-4 of Pathology. In A number of construction and new facilities will permit further Where Are They Now? 4 addition to the building renovation projects are expansion of cancer related re- Spotlight on Administration 5 annual influx of new residents, underway in the department and search in our department and help fellows and graduate students, several are nearing completion. I us to recruit additional outstanding New Changes in HIT 5-6 several new faculty members will am optimistic that by the time you clinicians and investigators to UAB. 6-7 From the Chief Residents be joining various divisions over read the next quarterly newsletter, From the Graduate Students 7-8 the next few months. Look for new the Anatomic Pathology Division faculty profiles in this and upcom- faculty, staff, and services cur- Best wishes, New Faculty Recruits 8 ing issues of our newsletter. Our rently housed in the Kracke Build- Kevin A. Roth, M.D., Ph.D. Pathology Grant Awards 9 aggressive recruitment strategy ing will all be moved to newly com- will continue through the fall as pleted facilities in PD6A. -

Bio-Tech/Life Science Industry Profile
ALABAMA’S BIO-INDUSTRY OVERVIEW Alabama’s multifaceted biosciences industry generates $7.3 billion in economic activity BIOSCIENCES annually while supporting 780 companies and nearly 48,000 direct and indirect jobs GENERATED across the state, according to an analysis by researchers at the University of Alabama. $7.3 BILLION Alabama’s biosciences sector is a robust combination of research institutions, labs, medical devices, innovative startups, pharmaceutical manufacturers, and more. IN ECONOMIC ACTIVITY BIO-INDUSTRY INNOVATION SUPPORTING Bioscience-Related U.S. Patents Bioscience-Related U.S. Patents 2014-2017 by Segment 2014-2017 780 53 Agricultural Chemicals COMPANIES 2014 161 Biochemistry 134 Bioinformatics & Health IT 9 2015 142 Biological Sampling & Analysis 42 Drugs & Pharmaceuticals 335 2016 159 Genetics 19 2017 141 Medical & Surgical Devices 240 Microbiology & Enzymes 37 Novel Plant Variants 14 Source: Bio.org, Alabama: Investment, Innovation and Job Creation in a Growing U.S. Bioscience Industry 2018 www.bio.org/value-bioscience-innovation-growing-jobs-and- improving-quality-life-2018 Bioscience Academic R&D Expenditures 48,000 $ Millions FY 2016 DIRECT AND Health Sciences $373.3 INDIRECTJOBS Biological & Biomedical $152.1 ACROSS THE Sciences Agricultural STATE Sciences $72.0 Other Life $29.4 Sciences Bioengineering Source: BioAlabama / Biomedical $4.0 Engineering www.edpa.org / (205)943-4700 / ALABAMA’S BIO-INDUSTRY NATIONAL INSTITUTES OF HEALTH FUNDING $450,000 NIH Funding ($ thousands) Alabama’s Top NIH-funded Research Institutions $400,000 $350,000 University of Alabama at Birmingham $300,000 University of South Alabama $250,000 Auburn University $200,000 University of Alabama $150,000 Southern Research Institute $100,000 $294,964 $298,159 $280,132 $228,361 $350,735 $391,578 $253,111 HudsonAlpha Institute of Biotechnology $50,000 $254,389 Source: National Institutes of Health, NIH Awards by Location & $- Organization - U.S. -

Peter Jeffrey Conn
Peter Jeffrey Conn 2806 Acklen Ave. Nashville, TN 37212 Professional Address P. Jeffrey Conn, Ph.D. Lee E. Limbird Professor of Pharmacology Director, Vanderbilt Center for Neuroscience Drug Discovery Department of Pharmacology Vanderbilt University 2215 Garland Avenue 1215-D Light Hall Nashville, TN 37232-6600 Phone: (615) 936-2478 Fax: (615) 343-3088 E-mail: [email protected] Education 1977 – 1981 B.S. Lee University, Cleveland, Tennessee (Psychology) (Magna Cum Laude) 1981 – 1982 University of Cincinnati, Cincinnati, Ohio (Physiological Psychology) 1982 – 1986 Ph.D. Vanderbilt University, Nashville, Tennessee (Pharmacology) Professional Training and Experience 1986 Research Associate, Department of Pharmacology, Vanderbilt University School of Medicine, Nashville, TN 1986 – 1988 Postdoctoral Fellow, Department of Pharmacology, Yale University School of Medicine, New Haven, CT 1988 Visiting Lecturer, Yale College, Yale University, New Haven, CT 1988 – 1994 Assistant Professor, Department of Pharmacology, Emory University School of Medicine, Atlanta, GA P. Jeffrey Conn – Curriculum Vitae Page 2 1994 – 1999 Associate Professor, Department of Pharmacology, Emory University School of Medicine, Atlanta, GA 1999 – 2000 Professor, Department of Pharmacology, Emory University School of Medicine, Atlanta, GA 2000 – 2003 Senior Director and Head, Department of Neuroscience, Merck Research Laboratories, Merck and Company, West Point, PA 2000 – 2005 Adjunct Professor, Department of Pharmacology, Emory University School of Medicine, Atlanta, -

Antibiotic and Antibacterial Vaccine Development Services
06/01/18 Antibiotic and Antibacterial Vaccine Development Services www.SouthernResearch.org/Drug-Development Contents Why choose Southern Research? ................................................................................................................. 4 in vitro Testing Services................................................................................................................................ 5 B0101 Minimum Inhibitory Concentration (MIC): ........................................................................................ 5 B0102 Minimum Bactericidal Concentration (MBC): .................................................................................... 5 B0103 Time-kill Assay: .................................................................................................................................. 5 B0104 Resistance Frequency Determination: ............................................................................................... 5 B0105 Post-antibiotic Effect (PAE): ............................................................................................................... 6 B0106 Serum and B0107 Protein Sensitivity: ............................................................................................... 6 B0108 Serum and B0109 Protein Binding: .................................................................................................... 6 B0110 Plasma and B0111 Microsome Stability: ........................................................................................... 6 B0112 -

Improved Orthopedic Surgical Frame
Improved Orthopedic Surgical Frame ALLIANCE FOR INNOVATIVE MEDICAL TECHNOLOGY – AIMTECH / TRAUMA Surgical retraction frame that can be quickly repositioned for procedures utilizing multiple surgical windows, especially for unstable pelvic fractures. Frame designed to be radio-lucent for use with X-ray procedures without needing to be disassembled or moved. ■■Technical Readiness Level ■■Inventors TRL 5 (Component validation in a relevant environment)— • Jason Lowe M.D., UAB Department Surgical frame made from carbon fiber components and of Orthopedic Surgery tested in a cadaver lab. • Alan Eberhardt, Ph.D., UAB Department ■■Competitive Advantages + Differentiation of Biomedical Engineering • Jason Lueck, Engineering, Southern Research Requires fewer surgical technicians to manually retract tissues, potentially reducing complications due to technician ■■Market Overview retraction error and fatigue. Reduced surgical time by Estimated 7M orthopedic fractures annually (US). Pelvic leaving frame in place during X-ray procedures. fractures represent 3%-6% of all fractures, and unstable ■■Intellectual Property Status fractures represent up to 20% of all pelvic fractures. ~63k. Patent Application WO 2016/611837 A1. PLEASE CONTACT Stacey Kelpke / Southern Research [email protected] / 205.581.2879 ABOUT SOUTHERN RESEARCH ABOUT UAB Founded in 1941 in Birmingham, Alabama, Southern Research is a scientific Known for its innovative and interdisciplinary approach to education at and engineering research organization that conducts preclinical drug both the graduate and undergraduate levels, the University of Alabama at discovery and development, advanced engineering research in materials, Birmingham is an internationally renowned research university and academic systems development, and energy and environmental technologies medical center, as well as Alabama’s largest employer, with some 23,000 research. -

Alabama's Medical Device Industry Overview
ALABAMA’S MEDICAL DEVICE INDUSTRY Baxter Incorporated makes OVERVIEW hemodialysis dialyzers, a high‐tech blood ‐filtering product used in kidney dialysis Alabama is known for its healthcare and biotechnology industries. The state has treatment. The facility is located in seven research universities, a highly accomplished contract research organization, a Opelika. biotechnology research institute, and dozens of life sciences companies. Access to talent, partnerships with higher education and industry, a low cost of living and BioHorizons, one of the fastest‐growing doing business creates a rich ecosystem for the medical device industry. global implant and biologics companies in the medical device industry, is located in Birmingham. Its porolio includes dental implants, instruments and biologics products. Southern Research (SR), a contract research organizaon with seven FDA‐ approved drugs on the market and campuses devoted to life science, engineering research and development, is located in Birmingham. Steris Corporaon has a global specialty HIGHLIGHTS services operaon headquartered in Birmingham. In 2014, Steris acquired IMS, a surgical device restoraon and Southern Research and the University of Alabama at Birmingham partnering through consulng company in Birmingham. the AIMTech project to develop life-changing medical devices. Over 600 bioscience companies, including 54 medical device and equipment Orchid Orthopedic Soluons is a world companies, employ approximately 1,600, according to U.S. Bureau of Labor Statistics leading medical device contract design data from 2014 Batelle/BIO report. and manufacturing company. The Average annual wage for the medical device and equipment industry is $55,958, facility, located in Arab, manufactures according to U.S. Bureau of Labor Statistics data from 2014 Batelle/BIO report. -

Tonix Pharmaceuticals and Southern Research Announce Expansion of COVID- 19 Vaccine Collaboration
June 18, 2020 Tonix Pharmaceuticals and Southern Research Announce Expansion of COVID- 19 Vaccine Collaboration Plan to Produce Blueprint of How the Human Immune System Responds to Infection Caused by SARS-CoV-2, the Virus that Causes COVID-19 Results Expected to Support Tonix’s Anticipated Regulatory Filings for TNX-1800, a Live Replicating Virus Vaccine Designed to Elicit T Cell Immunity in Addition to Antibodies NEW YORK, June 18, 2020 (GLOBE NEWSWIRE) -- Tonix Pharmaceuticals Holding Corp. (Nasdaq: TNXP) (Tonix or the Company), a clinical-stage biopharmaceutical company, announced today an expansion of its strategic collaboration with Southern Research to include a study of T cell immune responses to SARS-CoV-2 in volunteers who have recovered or remain asymptomatic after exposure to COVID-19. The research is part of an ongoing and broader collaboration between Tonix and Southern Research to develop and conduct animal testing of Tonix’s TNX-1800, which is a live replicating virus vaccine designed to protect against COVID-19. The data will support the interpretation of animal trial results with TNX-1800, which are expected in the fourth quarter of 2020 and subsequent human trials. “More than 200 years of vaccine experience, beginning with Dr. Edward Jenner’s landmark discoveries with horsepox and cowpox vaccines, have shown that T cell eliciting vaccines are particularly effective against viruses,” said Seth Lederman, M.D., President and CEO of Tonix. “We believe that protective vaccines against the SARS-CoV-2 virus will be similar in that regard. The data we plan to collect from recovered and asymptomatic COVID-19 volunteers will inform vaccine development on how to safely provide to vaccine recipients the same immune responses that others got from recovering from actual CoV-2 infection. -

National Council of University Research Administrat
Thank you! NCURA Region III wishes to thank our sponsors for their support! Corporate - Gold Level Corporate – Bronze Level Institutional 2 Program Committee Chair Jill Frazier Tincher, University of Miami Cynthia Hope, The University of Alabama Danielle McElwain, University of South Carolina Robyn Remotigue, Mississippi State University Riddick Smiley, East Carolina University Greg Thompson, Florida State University Mo Valentine, University of Arkansas for Medical Sciences Maria Valero-Martinez, University of Miami Michelle Vazin, Vanderbilt University Jeanne Ware, New College of Florida “Somewhere, something incredible is waiting to be known.” Carl Sagan 3 Welcome to NCURA Region III’s 2009 Meeting TABLE OF CONTENTS Page # Agenda 5 Sunday Schedule 8 Monday Schedule 14 Tuesday Schedule 22 Wednesday Schedule 32 Regional Committees 33 Hotel Maps 37 Program At-a-Glance 39 4 Agenda Tuesday, May 5, 2009 Continental Breakfast 7:30am – 9:00am Sunday, May 3, 2009 Morning Concurrent Sessions & Discussion Groups Registration 7:00am - 5:30pm 9:00am - 10:15am Continental Breakfast 7:00am - 8:30am Morning Break 10:15am - 10:30am Full-day Workshops Late Morning Concurrent Sessions 8:30am - 5:00pm & Discussion Groups Morning Workshops 10:30am - 11:45am 8:30am - 12:00pm Lunch 12:00pm - 1:00pm Morning Break 10:00am - 10:15am Afternoon Concurrent Sessions Lunch 12:00pm - 1:00pm 1:30pm - 2:45pm (Workshop participants only) Afternoon Break 2:45pm - 3:00pm Afternoon Workshops Late Afternoon Concurrent Sessions 1:30pm - 5:00pm & Discussion Groups Afternoon -

Nih Funding at Work in Alabama
Follow us on Twitter @UMR4NIH NIH FUNDING AT WORK IN ALABAMA QUICK FACTS NIH awarded $383.5 MILLION1 in grants and contracts during FY2020 that directly supported 6,008 JOBS2 and $922 MILLION2 economic activity in Alabama. The highest annual occurrences of disease deaths in Alabama are due to CANCER (10,632 lives lost), HEART DISEASE (13,473 lives lost) and STROKE (3,088 lives lost).3 Top-funded research institutions in Alabama include:1 University of Alabama at Birmingham Southern Research Institute Hudson-Alpha Institute for Biotechnology University of South Alabama Auburn University Jobs Supported by NIH Economic Activity Supported:2 Biopharmeceutical Industry Jobs:4 Biopharmeceutical Industry Extramural Research:2 Businesses:4 6,008 $922M 11,948 1,092 UMR is a coalition of leading research institutions, patient and health advocates and private industry seeking steady and sustainable increases in funding for the National Institutes of Health in order to save and improve lives, advance innovation and fuel the economy. The coalition members are: AdvaMed • Alzheimer's Association • American Association for the Advancement of Science • American Cancer Society Cancer Action Network • Association of American Universities • Association of Public and Land Grant Universities • BD • Boston University • Corning • Harvard University • Johns Hopkins University • Johnson & Johnson • Massachusetts Institute of Technology • Northwestern University • PhRMA • Stanford University • Thermo Fisher Scientific • University of Pennsylvania • Vanderbilt University • Vanderbilt University Medical Center • Washington University in St. Louis 1. National Institutes of Health (NIH), NIH Awards by Location & Organization, U.S. FY2020 2. United for Medical Research Original Data - U.S. FY2020 3. Centers for Disease Control, "National Vital Statistics Report: Deaths: Final Data for 2018" Page 58. -
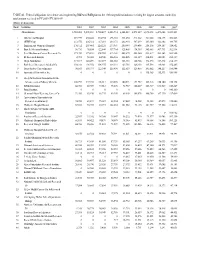
TABLE 46. Federal Obligations for Science and Engineering R&D And
TABLE 46. Federal obligations for science and engineering R&D and R&D plant to the 100 nonprofit institutions receiving the largest amounts, ranked by total amount received in FY 2009: FY 2001–09 (Dollars in thousands) Rank Institution 2001 2002 2003 2004 2005 2006 2007 2008 2009a All institutions 3,933,088 5,278,301 5,720,427 6,083,714 6,243,849 6,572,639 6,872,498 6,471,044 8,321,301 1 MA General Hospital 219,979 250,440 294,794 296,765 301,630 311,202 321,660 316,173 382,563 2 MITRE Corp. 212,576 226,312 187,210 284,478 282,943 307,239 305,360 322,882 344,765 3 Brigham and Women's Hospital 176,312 207,465 220,221 233,907 250,949 239,456 258,176 254,107 324,852 4 Battelle Memorial Institute 36,710 98,854 122,844 137,764 125,405 194,989 345,463 307,733 302,914 5 Fred Hutchinson Cancer Research Ctr. 173,490 175,631 208,055 213,461 202,272 253,460 231,817 224,163 263,980 6 IIT Research Institute 2,783 58,020 64,948 126,516 118,405 131,519 134,031 142,003 232,359 7 Mayo Foundation 117,017 145,499 163,077 208,152 181,118 188,966 192,570 187,372 214,139 8 Beth Israel Deaconess Medical Ctr. 103,331 101,072 106,555 114,331 125,781 126,403 115,594 114,981 152,205 9 Dana-Farber Cancer Institute 96,563 103,973 122,344 125,490 112,519 128,455 141,002 140,217 150,944 10 Associated Universities Inc. -

Chapter 9 Growing Economic Base Industries $395K
$500K Chapter 9 $395K Growing Economic Base Industries $300K $200K $100K PERSONAL VISION STATEMENTS $33K “Having a culture and tools 2000 2001 2002 2003 2004 2005 2006 2007 2008 2009 2010 2011 2012 2-BEDROOM 3-BEDROOM 4-BEDROOM ALL PROPERTIES that allow for innovation and entrepreneurship.” “Economic stability for residents, businesses, and persons looking to do business in the city...” 9.1 CITY OF BIRMINGHAM COMPREHENSIVE PLAN PART IV | CHAPTER 9 GROWING ECONOMIC BASE INDUSTRIES GOALS POLICIES FOR DECISION MAKERS A diversified, knowledge-driven economy, • Target city resources and support to existing and emerging economic base anchored by established and emerging industries. industry sectors, built on the city’s competitive strengths, and providing job • Support the development of a diverse economy that provides a wide range of opportunities in a range of occupations employment opportunities and shields the city from over-dependence on any single and skill levels. industry. • Leverage city resources through collaboration with key partners in the private and nonprofit sectors. • Engage with the business community to ensure that city policies are responsive to business needs. • Advocate for state policies and investments that support the city’s economic development. • Collaborate at the regional level when regional action is required to advance the city’s economic interests. A city and its government that are focused • Strive to create and maintain an award-winning educational system, both in the on economic development. Pre-K and K-12 systems and in higher education. • Ensure that the City’s workforce development programs are meeting the needs of its citizens and of current and future employers.