Characterization of Bacterial Diversity in Cold-Water Anthothelidae Corals" (2016)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genomic Insight Into the Host–Endosymbiont Relationship of Endozoicomonas Montiporae CL-33T with Its Coral Host
ORIGINAL RESEARCH published: 08 March 2016 doi: 10.3389/fmicb.2016.00251 Genomic Insight into the Host–Endosymbiont Relationship of Endozoicomonas montiporae CL-33T with its Coral Host Jiun-Yan Ding 1, Jia-Ho Shiu 1, Wen-Ming Chen 2, Yin-Ru Chiang 1 and Sen-Lin Tang 1* 1 Biodiversity Research Center, Academia Sinica, Taipei, Taiwan, 2 Department of Seafood Science, Laboratory of Microbiology, National Kaohsiung Marine University, Kaohsiung, Taiwan The bacterial genus Endozoicomonas was commonly detected in healthy corals in many coral-associated bacteria studies in the past decade. Although, it is likely to be a core member of coral microbiota, little is known about its ecological roles. To decipher potential interactions between bacteria and their coral hosts, we sequenced and investigated the first culturable endozoicomonal bacterium from coral, the E. montiporae CL-33T. Its genome had potential sign of ongoing genome erosion and gene exchange with its Edited by: Rekha Seshadri, host. Testosterone degradation and type III secretion system are commonly present in Department of Energy Joint Genome Endozoicomonas and may have roles to recognize and deliver effectors to their hosts. Institute, USA Moreover, genes of eukaryotic ephrin ligand B2 are present in its genome; presumably, Reviewed by: this bacterium could move into coral cells via endocytosis after binding to coral’s Eph Kathleen M. Morrow, University of New Hampshire, USA receptors. In addition, 7,8-dihydro-8-oxoguanine triphosphatase and isocitrate lyase Jean-Baptiste Raina, are possible type III secretion effectors that might help coral to prevent mitochondrial University of Technology Sydney, Australia dysfunction and promote gluconeogenesis, especially under stress conditions. -

Coelenterata: Anthozoa), with Diagnoses of New Taxa
PROC. BIOL. SOC. WASH. 94(3), 1981, pp. 902-947 KEY TO THE GENERA OF OCTOCORALLIA EXCLUSIVE OF PENNATULACEA (COELENTERATA: ANTHOZOA), WITH DIAGNOSES OF NEW TAXA Frederick M. Bayer Abstract.—A serial key to the genera of Octocorallia exclusive of the Pennatulacea is presented. New taxa introduced are Olindagorgia, new genus for Pseudopterogorgia marcgravii Bayer; Nicaule, new genus for N. crucifera, new species; and Lytreia, new genus for Thesea plana Deich- mann. Ideogorgia is proposed as a replacement ñame for Dendrogorgia Simpson, 1910, not Duchassaing, 1870, and Helicogorgia for Hicksonella Simpson, December 1910, not Nutting, May 1910. A revised classification is provided. Introduction The key presented here was an essential outgrowth of work on a general revisión of the octocoral fauna of the western part of the Atlantic Ocean. The far-reaching zoogeographical affinities of this fauna made it impossible in the course of this study to ignore genera from any part of the world, and it soon became clear that many of them require redefinition according to modern taxonomic standards. Therefore, the type-species of as many genera as possible have been examined, often on the basis of original type material, and a fully illustrated generic revisión is in course of preparation as an essential first stage in the redescription of western Atlantic species. The key prepared to accompany this generic review has now reached a stage that would benefit from a broader and more objective testing under practical conditions than is possible in one laboratory. For this reason, and in order to make the results of this long-term study available, even in provisional form, not only to specialists but also to the growing number of ecologists, biochemists, and physiologists interested in octocorals, the key is now pre- sented in condensed form with minimal illustration. -

St. Kitts Final Report
ReefFix: An Integrated Coastal Zone Management (ICZM) Ecosystem Services Valuation and Capacity Building Project for the Caribbean ST. KITTS AND NEVIS FIRST DRAFT REPORT JUNE 2013 PREPARED BY PATRICK I. WILLIAMS CONSULTANT CLEVERLY HILL SANDY POINT ST. KITTS PHONE: 1 (869) 765-3988 E-MAIL: [email protected] 1 2 TABLE OF CONTENTS Page No. Table of Contents 3 List of Figures 6 List of Tables 6 Glossary of Terms 7 Acronyms 10 Executive Summary 12 Part 1: Situational analysis 15 1.1 Introduction 15 1.2 Physical attributes 16 1.2.1 Location 16 1.2.2 Area 16 1.2.3 Physical landscape 16 1.2.4 Coastal zone management 17 1.2.5 Vulnerability of coastal transportation system 19 1.2.6 Climate 19 1.3 Socio-economic context 20 1.3.1 Population 20 1.3.2 General economy 20 1.3.3 Poverty 22 1.4 Policy frameworks of relevance to marine resource protection and management in St. Kitts and Nevis 23 1.4.1 National Environmental Action Plan (NEAP) 23 1.4.2 National Physical Development Plan (2006) 23 1.4.3 National Environmental Management Strategy (NEMS) 23 1.4.4 National Biodiversity Strategy and Action Plan (NABSAP) 26 1.4.5 Medium Term Economic Strategy Paper (MTESP) 26 1.5 Legislative instruments of relevance to marine protection and management in St. Kitts and Nevis 27 1.5.1 Development Control and Planning Act (DCPA), 2000 27 1.5.2 National Conservation and Environmental Protection Act (NCEPA), 1987 27 1.5.3 Public Health Act (1969) 28 1.5.4 Solid Waste Management Corporation Act (1996) 29 1.5.5 Water Courses and Water Works Ordinance (Cap. -

Preliminary Report on the Octocorals (Cnidaria: Anthozoa: Octocorallia) from the Ogasawara Islands
国立科博専報,(52), pp. 65–94 , 2018 年 3 月 28 日 Mem. Natl. Mus. Nat. Sci., Tokyo, (52), pp. 65–94, March 28, 2018 Preliminary Report on the Octocorals (Cnidaria: Anthozoa: Octocorallia) from the Ogasawara Islands Yukimitsu Imahara1* and Hiroshi Namikawa2 1Wakayama Laboratory, Biological Institute on Kuroshio, 300–11 Kire, Wakayama, Wakayama 640–0351, Japan *E-mail: [email protected] 2Showa Memorial Institute, National Museum of Nature and Science, 4–1–1 Amakubo, Tsukuba, Ibaraki 305–0005, Japan Abstract. Approximately 400 octocoral specimens were collected from the Ogasawara Islands by SCUBA diving during 2013–2016 and by dredging surveys by the R/V Koyo of the Tokyo Met- ropolitan Ogasawara Fisheries Center in 2014 as part of the project “Biological Properties of Bio- diversity Hotspots in Japan” at the National Museum of Nature and Science. Here we report on 52 lots of these octocoral specimens that have been identified to 42 species thus far. The specimens include seven species of three genera in two families of Stolonifera, 25 species of ten genera in two families of Alcyoniina, one species of Scleraxonia, and nine species of four genera in three families of Pennatulacea. Among them, three species of Stolonifera: Clavularia cf. durum Hick- son, C. cf. margaritiferae Thomson & Henderson and C. cf. repens Thomson & Henderson, and five species of Alcyoniina: Lobophytum variatum Tixier-Durivault, L. cf. mirabile Tixier- Durivault, Lohowia koosi Alderslade, Sarcophyton cf. boletiforme Tixier-Durivault and Sinularia linnei Ofwegen, are new to Japan. In particular, Lohowia koosi is the first discovery since the orig- inal description from the east coast of Australia. -

Endozoicomonas Are Specific, Facultative Symbionts of Sea Squirts
ORIGINAL RESEARCH published: 12 July 2016 doi: 10.3389/fmicb.2016.01042 Endozoicomonas Are Specific, Facultative Symbionts of Sea Squirts Lars Schreiber 1*, Kasper U. Kjeldsen 1, Peter Funch 2, Jeppe Jensen 1, Matthias Obst 3, Susanna López-Legentil 4 and Andreas Schramm 1 1 Department of Bioscience, Center for Geomicrobiology and Section for Microbiology, Aarhus University, Aarhus, Denmark, 2 Section of Genetics, Ecology and Evolution, Department of Bioscience, Aarhus University, Aarhus, Denmark, 3 Department of Marine Sciences, University of Gothenburg, Gothenburg, Sweden, 4 Department of Biology and Marine Biology, Center for Marine Science, University of North Carolina Wilmington, Wilmington NC, USA Ascidians are marine filter feeders and harbor diverse microbiota that can exhibit a high degree of host-specificity. Pharyngeal samples of Scandinavian and Mediterranean ascidians were screened for consistently associated bacteria by culture-dependent and -independent approaches. Representatives of the Endozoicomonas (Gammaproteobacteria, Hahellaceae) clade were detected in the ascidian species Ascidiella aspersa, Ascidiella scabra, Botryllus schlosseri, Ciona intestinalis, Styela clava, and multiple Ascidia/Ascidiella spp. In total, Endozoicomonas was detected in more than half of all specimens screened, and in 25–100% of the specimens for each species. The retrieved Endozoicomonas 16S rRNA gene sequences formed an ascidian-specific subclade, whose members were detected by fluorescence Edited by: in situ hybridization (FISH) as extracellular microcolonies in the pharynx. Two strains Joerg Graf, of the ascidian-specific Endozoicomonas subclade were isolated in pure culture and University of Connecticut, USA characterized. Both strains are chemoorganoheterotrophs and grow on mucin (a Reviewed by: Silvia Bulgheresi, mucus glycoprotein). The strains tested negative for cytotoxic or antibacterial activity. -

Marine Ecology Progress Series 479:75–84 (2013)
The following supplement accompanies the article Bacteria of the genus Endozoicomonas dominate the microbiome of the Mediterranean gorgonian coral Eunicella cavolini Till Bayer1,*, Chatchanit Arif1, Christine Ferrier-Pagès2, Didier Zoccola2, Manuel Aranda1, Christian R. Voolstra1,* 1Red Sea Research Center, King Abdullah University of Science and Technology, 23955 Thuwal, Kingdom of Saudi Arabia 2Centre Scientifique de Monaco, 98000 Monaco, Monaco *Corresponding authors. Emails: [email protected] and [email protected] Marine Ecology Progress Series 479:75–84 (2013) Supplement. Data supplementing the alpha and beta diversity measurements, as well as details of the sample-group to OTU associations W41 W24 1500 W30 1000 G24A Number of O TUs Number of O G30A 500 G41C G24C G30C G30B G41A G41B G24B 0 0 10000 20000 30000 Number of sequences Fig. S1. Rarefaction curves for all samples. Curves show the number of OTUs at different sampling depths. Fig. S2. Principal coordinate analysis of weighted UniFrac distances. The percentages of variability explained by each axis are given in parentheses. 2 Fig. S3. Phylogenetic tree of the Encozoicomonas from Eunicella cavolini (blue) and closely related bacteria. Most bacteria represented originate from marine invertebrates, such as soft corals (Alcyonacea, red) or scleractinian corals (green). Others include bivalves, sponges or sea slugs. EU884929 represents a bacterium from a fish, Pomacanthus sextriatus. Type strains are marked with *. The tree was calculated using PhyML as implemented in ARB, with a GTR substitution model. Bootstrap values are shown for 1000 bootsraps of PhyML, and 1000 bootstraps of a neighborhood joining tree calculated in MEGA 5. Full length 16S sequences obtained from cloning were used for this tree. -
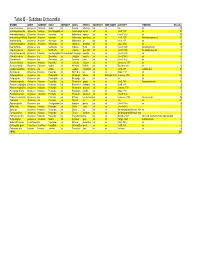
Table B – Subclass Octocorallia
Table B – Subclass Octocorallia BINOMEN ORDER SUBORDER FAMILY SUBFAMILY GENUS SPECIES SUBSPECIES COMN_NAMES AUTHORITY SYNONYMS #Records Acanella arbuscula Alcyonacea Calcaxonia Isididae n/a Acanella arbuscula n/a n/a n/a n/a 59 Acanthogorgia armata Alcyonacea Holaxonia Acanthogorgiidae n/a Acanthogorgia armata n/a n/a Verrill, 1878 n/a 95 Anthomastus agassizii Alcyonacea Alcyoniina Alcyoniidae n/a Anthomastus agassizii n/a n/a (Verrill, 1922) n/a 35 Anthomastus grandiflorus Alcyonacea Alcyoniina Alcyoniidae n/a Anthomastus grandiflorus n/a n/a Verrill, 1878 Anthomastus purpureus 37 Anthomastus sp. Alcyonacea Alcyoniina Alcyoniidae n/a Anthomastus sp. n/a n/a Verrill, 1878 n/a 1 Anthothela grandiflora Alcyonacea Scleraxonia Anthothelidae n/a Anthothela grandiflora n/a n/a (Sars, 1856) n/a 24 Capnella florida Alcyonacea n/a Nephtheidae n/a Capnella florida n/a n/a (Verrill, 1869) Eunephthya florida 44 Capnella glomerata Alcyonacea n/a Nephtheidae n/a Capnella glomerata n/a n/a (Verrill, 1869) Eunephthya glomerata 4 Chrysogorgia agassizii Alcyonacea Holaxonia Acanthogorgiidae Chrysogorgiidae Chrysogorgia agassizii n/a n/a (Verrill, 1883) n/a 2 Clavularia modesta Alcyonacea n/a Clavulariidae n/a Clavularia modesta n/a n/a (Verrill, 1987) n/a 6 Clavularia rudis Alcyonacea n/a Clavulariidae n/a Clavularia rudis n/a n/a (Verrill, 1922) n/a 1 Gersemia fruticosa Alcyonacea Alcyoniina Alcyoniidae n/a Gersemia fruticosa n/a n/a Marenzeller, 1877 n/a 3 Keratoisis flexibilis Alcyonacea Calcaxonia Isididae n/a Keratoisis flexibilis n/a n/a Pourtales, 1868 n/a 1 Lepidisis caryophyllia Alcyonacea n/a Isididae n/a Lepidisis caryophyllia n/a n/a Verrill, 1883 Lepidisis vitrea 13 Muriceides sp. -

Supporting Information
Supporting Information Lozupone et al. 10.1073/pnas.0807339105 SI Methods nococcus, and Eubacterium grouped with members of other Determining the Environmental Distribution of Sequenced Genomes. named genera with high bootstrap support (Fig. 1A). One To obtain information on the lifestyle of the isolate and its reported member of the Bacteroidetes (Bacteroides capillosus) source, we looked at descriptive information from NCBI grouped firmly within the Firmicutes. This taxonomic error was (www.ncbi.nlm.nih.gov/genomes/lproks.cgi) and other related not surprising because gut isolates have often been classified as publications. We also determined which 16S rRNA-based envi- Bacteroides based on an obligate anaerobe, Gram-negative, ronmental surveys of microbial assemblages deposited near- nonsporulating phenotype alone (6, 7). A more recent 16S identical sequences in GenBank. We first downloaded the gbenv rRNA-based analysis of the genus Clostridium defined phylo- files from the NCBI ftp site on December 31, 2007, and used genetically related clusters (4, 5), and these designations were them to create a BLAST database. These files contain GenBank supported in our phylogenetic analysis of the Clostridium species in the HGMI pipeline. We thus designated these Clostridium records for the ENV database, a component of the nonredun- species, along with the species from other named genera that dant nucleotide database (nt) where 16S rRNA environmental cluster with them in bootstrap supported nodes, as being within survey data are deposited. GenBank records for hits with Ͼ98% these clusters. sequence identity over 400 bp to the 16S rRNA sequence of each of the 67 genomes were parsed to get a list of study titles Annotation of GTs and GHs. -

Deep‐Sea Coral Taxa in the U.S. Gulf of Mexico: Depth and Geographical Distribution
Deep‐Sea Coral Taxa in the U.S. Gulf of Mexico: Depth and Geographical Distribution by Peter J. Etnoyer1 and Stephen D. Cairns2 1. NOAA Center for Coastal Monitoring and Assessment, National Centers for Coastal Ocean Science, Charleston, SC 2. National Museum of Natural History, Smithsonian Institution, Washington, DC This annex to the U.S. Gulf of Mexico chapter in “The State of Deep‐Sea Coral Ecosystems of the United States” provides a list of deep‐sea coral taxa in the Phylum Cnidaria, Classes Anthozoa and Hydrozoa, known to occur in the waters of the Gulf of Mexico (Figure 1). Deep‐sea corals are defined as azooxanthellate, heterotrophic coral species occurring in waters 50 m deep or more. Details are provided on the vertical and geographic extent of each species (Table 1). This list is adapted from species lists presented in ʺBiodiversity of the Gulf of Mexicoʺ (Felder & Camp 2009), which inventoried species found throughout the entire Gulf of Mexico including areas outside U.S. waters. Taxonomic names are generally those currently accepted in the World Register of Marine Species (WoRMS), and are arranged by order, and alphabetically within order by suborder (if applicable), family, genus, and species. Data sources (references) listed are those principally used to establish geographic and depth distribution. Only those species found within the U.S. Gulf of Mexico Exclusive Economic Zone are presented here. Information from recent studies that have expanded the known range of species into the U.S. Gulf of Mexico have been included. The total number of species of deep‐sea corals documented for the U.S. -

The Genetic Identity of Dinoflagellate Symbionts in Caribbean Octocorals
Coral Reefs (2004) 23: 465-472 DOI 10.1007/S00338-004-0408-8 REPORT Tamar L. Goulet • Mary Alice CofFroth The genetic identity of dinoflagellate symbionts in Caribbean octocorals Received: 2 September 2002 / Accepted: 20 December 2003 / Published online: 29 July 2004 © Springer-Verlag 2004 Abstract Many cnidarians (e.g., corals, octocorals, sea Introduction anemones) maintain a symbiosis with dinoflagellates (zooxanthellae). Zooxanthellae are grouped into The cornerstone of the coral reef ecosystem is the sym- clades, with studies focusing on scleractinian corals. biosis between cnidarians (e.g., corals, octocorals, sea We characterized zooxanthellae in 35 species of Caribbean octocorals. Most Caribbean octocoral spe- anemones) and unicellular dinoñagellates commonly called zooxanthellae. Studies of zooxanthella symbioses cies (88.6%) hosted clade B zooxanthellae, 8.6% have previously been hampered by the difficulty of hosted clade C, and one species (2.9%) hosted clades B and C. Erythropodium caribaeorum harbored clade identifying the algae. Past techniques relied on culturing and/or identifying zooxanthellae based on their free- C and a unique RFLP pattern, which, when se- swimming form (Trench 1997), antigenic features quenced, fell within clade C. Five octocoral species (Kinzie and Chee 1982), and cell architecture (Blank displayed no zooxanthella cladal variation with depth. 1987), among others. These techniques were time-con- Nine of the ten octocoral species sampled throughout suming, required a great deal of expertise, and resulted the Caribbean exhibited no regional zooxanthella cla- in the differentiation of only a small number of zoo- dal differences. The exception, Briareum asbestinum, xanthella species. Molecular techniques amplifying had some colonies from the Dry Tortugas exhibiting zooxanthella DNA encoding for the small and large the E. -

Deep-Sea Coral Taxa in the U. S. Caribbean Region: Depth And
Deep‐Sea Coral Taxa in the U. S. Caribbean Region: Depth and Geographical Distribution By Stephen D. Cairns1 1. National Museum of Natural History, Smithsonian Institution, Washington, DC An update of the status of the azooxanthellate, heterotrophic coral species that occur predominantly deeper than 50 m in the U.S. Caribbean territories is not given in this volume because of lack of significant additional data. However, an updated list of deep‐sea coral species in Phylum Cnidaria, Classes Anthozoa and Hydrozoa, from the Caribbean region (Figure 1) is presented below. Details are provided on depth ranges and known geographic distributions within the region (Table 1). This list is adapted from Lutz & Ginsburg (2007, Appendix 8.1) in that it is restricted to the U. S. territories in the Caribbean, i.e., Puerto Rico, U. S. Virgin Islands, and Navassa Island, not the entire Caribbean and Bahamian region. Thus, this list is significantly shorter. The list has also been reordered alphabetically by family, rather than species, to be consistent with other regional lists in this volume, and authorship and publication dates have been added. Also, Antipathes americana is now properly assigned to the genus Stylopathes, and Stylaster profundus to the genus Stenohelia. Furthermore, many of the geographic ranges have been clarified and validated. Since 2007 there have been 20 species additions to the U.S. territories list (indicated with blue shading in the list), mostly due to unpublished specimens from NMNH collections. As a result of this update, there are now known to be: 12 species of Antipatharia, 45 species of Scleractinia, 47 species of Octocorallia (three with incomplete taxonomy), and 14 species of Stylasteridae, for a total of 118 species found in the relatively small geographic region of U. -

Volume III of This Document)
4.1.3 Coastal Migratory Pelagics Description and Distribution (from CMP Am 15) The coastal migratory pelagics management unit includes cero (Scomberomous regalis), cobia (Rachycentron canadum), king mackerel (Scomberomous cavalla), Spanish mackerel (Scomberomorus maculatus) and little tunny (Euthynnus alleterattus). The mackerels and tuna in this management unit are often referred to as ―scombrids.‖ The family Scombridae includes tunas, mackerels and bonitos. They are among the most important commercial and sport fishes. The habitat of adults in the coastal pelagic management unit is the coastal waters out to the edge of the continental shelf in the Atlantic Ocean. Within the area, the occurrence of coastal migratory pelagic species is governed by temperature and salinity. All species are seldom found in water temperatures less than 20°C. Salinity preference varies, but these species generally prefer high salinity. The scombrids prefer high salinities, but less than 36 ppt. Salinity preference of little tunny and cobia is not well defined. The larval habitat of all species in the coastal pelagic management unit is the water column. Within the spawning area, eggs and larvae are concentrated in the surface waters. (from PH draft Mackerel Am. 18) King Mackerel King mackerel is a marine pelagic species that is found throughout the Gulf of Mexico and Caribbean Sea and along the western Atlantic from the Gulf of Maine to Brazil and from the shore to 200 meter depths. Adults are known to spawn in areas of low turbidity, with salinity and temperatures of approximately 30 ppt and 27°C, respectively. There are major spawning areas off Louisiana and Texas in the Gulf (McEachran and Finucane 1979); and off the Carolinas, Cape Canaveral, and Miami in the western Atlantic (Wollam 1970; Schekter 1971; Mayo 1973).