IR and Raman Spectroscopy
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Comparison of Ultraviolet and Visible Raman Spectra of Supported Metal Oxide Catalysts
8600 J. Phys. Chem. B 2001, 105, 8600-8606 A Comparison of Ultraviolet and Visible Raman Spectra of Supported Metal Oxide Catalysts Yek Tann Chua,† Peter C. Stair,*,† and Israel E. Wachs*,‡ Department of Chemistry, Center for Catalysis and Surface Science and Institute of EnVironmental Catalysis, Northwestern UniVersity, EVanston, Illinois 60208, and Zettlemoyer Center for Surface Studies and Department of Chemical Engineering, Lehigh UniVersity, Bethlehem, PennsylVania 18015 ReceiVed: April 11, 2001 The recent emergence of ultraviolet-wavelength-excited Raman spectroscopy as a tool for catalyst characterization has motivated the question of how UV Raman spectra compare to visible-wavelength-excited Raman spectra on the same catalyst system. Measurements of Raman spectra from five supported metal oxide systems (Al2O3-supported Cr2O3,V2O5, and MoO3 as well as TiO2-supported MoO3 and Re2O7), using visible (514.5 nm) and ultraviolet (244 nm) wavelength excitation have been compared to determine the similarities and differences in Raman spectra produced at the two wavelengths. The samples were in the form of self-supporting disks. Spectra from the oxides, both hydrated as a result of contact with ambient air and dehydrated as a result of calcination or laser-induced heating, were recorded. A combination of sample spinning and translation to produce a spiral pattern of laser beam exposure to the catalyst disk was found to be most effective in minimizing dehydration caused by laser-induced heating. Strong absorption by the samples in the ultraviolet significantly reduced the number of scatterers contributing to the Raman spectrum while producing only modest increases in the Raman scattering cross section due to resonance enhancement. -

Atomic and Molecular Laser-Induced Breakdown Spectroscopy of Selected Pharmaceuticals
Article Atomic and Molecular Laser-Induced Breakdown Spectroscopy of Selected Pharmaceuticals Pravin Kumar Tiwari 1,2, Nilesh Kumar Rai 3, Rohit Kumar 3, Christian G. Parigger 4 and Awadhesh Kumar Rai 2,* 1 Institute for Plasma Research, Gandhinagar, Gujarat-382428, India 2 Laser Spectroscopy Research Laboratory, Department of Physics, University of Allahabad, Prayagraj-211002, India 3 CMP Degree College, Department of Physics, University of Allahabad, Pragyagraj-211002, India 4 Physics and Astronomy Department, University of Tennessee, University of Tennessee Space Institute, Center for Laser Applications, 411 B.H. Goethert Parkway, Tullahoma, TN 37388-9700, USA * Correspondence: [email protected]; Tel.: +91-532-2460993 Received: 10 June 2019; Accepted: 10 July 2019; Published: 19 July 2019 Abstract: Laser-induced breakdown spectroscopy (LIBS) of pharmaceutical drugs that contain paracetamol was investigated in air and argon atmospheres. The characteristic neutral and ionic spectral lines of various elements and molecular signatures of CN violet and C2 Swan band systems were observed. The relative hardness of all drug samples was measured as well. Principal component analysis, a multivariate method, was applied in the data analysis for demarcation purposes of the drug samples. The CN violet and C2 Swan spectral radiances were investigated for evaluation of a possible correlation of the chemical and molecular structures of the pharmaceuticals. Complementary Raman and Fourier-transform-infrared spectroscopies were used to record the molecular spectra of the drug samples. The application of the above techniques for drug screening are important for the identification and mitigation of drugs that contain additives that may cause adverse side-effects. Keywords: paracetamol; laser-induced breakdown spectroscopy; cyanide; carbon swan bands; principal component analysis; Raman spectroscopy; Fourier-transform-infrared spectroscopy 1. -

Laser Raman Spectroscopy As a Technique for Identification Of
ARTICLE IN PRESS CHEMGE-15589; No of Pages 13 Chemical Geology xxx (2008) xxx–xxx Contents lists available at ScienceDirect Chemical Geology journal homepage: www.elsevier.com/locate/chemgeo Laser Raman spectroscopy as a technique for identification of seafloor hydrothermal and cold seep minerals Sheri N. White ⁎ Department of Applied Ocean Physics and Engineering, Woods Hole Oceanographic Institution, Woods Hole, MA 02536, USA article info abstract Article history: In situ sensors capable of real-time measurements and analyses in the deep ocean are necessary to fulfill the Received 8 August 2008 potential created by the development of autonomous, deep-sea platforms such as autonomous and remotely Received in revised form 8 November 2008 operated vehicles, and cabled observatories. Laser Raman spectroscopy (a type of vibrational spectroscopy) is an Accepted 10 November 2008 optical technique that is capable of in situ molecular identification of minerals in the deep ocean. The goals of this Available online xxxx work are to determine the characteristic spectral bands and relative Raman scattering strength of hydrothermally- Editor: R.L. Rudnick and cold seep-relevant minerals, and to determine how the quality of the spectra are affected by changes in excitation wavelength and sampling optics. The information learned from this work will lead to the development Keywords: of new, smaller sea-going Raman instruments that are optimized to analyze minerals in the deep ocean. Raman spectroscopy Many minerals of interest at seafloor hydrothermal and cold seep sites are Raman active, such as elemental sulfur, Mineralogy carbonates, sulfates and sulfides. Elemental S8 sulfur is a strong Raman scatterer with dominant bands at ∼219 and Hydrothermal vents 472 Δcm−1. -

Using Visible and Near-Infrared Reflectance Spectroscopy and Differential Scanning Calorimetry to Study Starch, Protein, and Temperature Effects on Bread Staling
Using Visible and Near-Infrared Reflectance Spectroscopy and Differential Scanning Calorimetry to Study Starch, Protein, and Temperature Effects on Bread Staling Feng Xie,1 Floyd E. Dowell,2,3 and Xiuzhi S. Sun1 ABSTRACT Cereal Chem. 81(2):249–254 Starch, protein, and temperature effects on bread staling were inves- retrograded amylose-lipid complex was limited. Two important wave- tigated using visible and near-infrared spectroscopy (NIRS) and differ- lengths, 550 nm and 1,465 nm, were key for NIRS to successfully classify ential scanning calorimetry (DSC). Bread staling was mainly due to the starch-starch (SS) and starch-protein (SP) bread based on different amylopectin retrogradation. NIRS measured amylopectin retrogradation colors and protein contents in SS and SP. Low temperature dramatically accurately in different batches. Three important wavelengths, 970 nm, accelerated the amylopectin retrogradation process. Protein retarded bread 1,155 nm, and 1,395 nm, were associated with amylopectin retrogra- staling, but not as much as temperature. The starch and protein interaction dation. NIRS followed moisture and starch structure changes when was less important than the starch retrogradation. Protein hindered the amylopectin retrograded. The amylose-lipid complex changed little from bread staling process mainly by diluting starch and retarding starch retro- one day after baking. The capability of NIRS to measure changes in the gradation. Bread staling is a complex process that occurs during bread and starch (Osborne and Douglas 1981; Davies and Miller 1988; storage. It is a progressive deterioration of qualities such as taste, Millar et al 1996). Therefore, NIRS has the potential to provide fun- firmness, etc. -

Accessing Excited State Molecular Vibrations by Femtosecond Stimulated Raman Spectroscopy
Accessing Excited State Molecular Vibrations by Femtosecond Stimulated Raman Spectroscopy Giovanni Batignani,y Carino Ferrante,y,z and Tullio Scopigno∗,y,z yDipartimento di Fisica, Universitá di Roma “La Sapienza", Roma, I-00185, Italy z Istituto Italiano di Tecnologia, Center for Life Nano Science @Sapienza, Roma, I-00161, Italy E-mail: [email protected] arXiv:2010.05029v1 [physics.optics] 10 Oct 2020 1 Abstract Excited-state vibrations are crucial for determining photophysical and photochem- ical properties of molecular compounds. Stimulated Raman scattering can coherently stimulate and probe molecular vibrations with optical pulses, but it is generally re- stricted to ground state properties. Working in resonance conditions, indeed, enables cross-section enhancement and selective excitation to a targeted electronic level, but is hampered by an increased signal complexity due to the presence of overlapping spectral contributions. Here, we show how detailed information on ground and excited state vi- brations can be disentangled, by exploiting the relative time delay between Raman and probe pulses to control the excited state population, combined with a diagrammatic formalism to dissect the pathways concurring to the signal generation. The proposed method is then exploited to elucidate the vibrational properties of ground and excited electronic states in the paradigmatic case of Cresyl Violet. We anticipate that the presented approach holds the potential for selective mapping the reaction coordinates pertaining to transient electronic stages implied in photo-active compounds. Graphical TOC Entry 2 Raman spectroscopy is a powerful tool to access the vibrational fingerprints of molecules or solid state compounds and it can be used to extract structural and dynamical information of the samples under investigation. -

Combining Chemical Information from Grass Pollen in Multimodal Characterization
ORIGINAL RESEARCH published: 31 January 2020 doi: 10.3389/fpls.2019.01788 Combining Chemical Information From Grass Pollen in Multimodal Characterization Sabrina Diehn 1,2, Boris Zimmermann 3, Valeria Tafintseva 3, Stephan Seifert 1,2, Murat Bag˘ cıog˘ lu 3, Mikael Ohlson 4, Steffen Weidner 2, Siri Fjellheim 5, Achim Kohler 3,6 and Janina Kneipp 1,2* 1 Department of Chemistry, Humboldt-Universität zu Berlin, Berlin, Germany, 2 BAM Federal Institute for Materials Research and Testing, Berlin, Germany, 3 Faculty of Science and Technology, Norwegian University of Life Sciences, Ås, Norway, 4 Faculty of Environmental Sciences and Natural Resource Management, Norwegian University of Life Sciences, Ås, Norway, 5 Faculty of Biosciences, Norwegian University of Life Sciences, Ås, Norway, 6 Nofima AS, Ås, Norway Edited by: Lisbeth Garbrecht Thygesen, fi University of Copenhagen, The analysis of pollen chemical composition is important to many elds, including Denmark agriculture, plant physiology, ecology, allergology, and climate studies. Here, the Reviewed by: potential of a combination of different spectroscopic and spectrometric methods Wesley Toby Fraser, regarding the characterization of small biochemical differences between pollen samples Oxford Brookes University, United Kingdom was evaluated using multivariate statistical approaches. Pollen samples, collected from Åsmund Rinnan, three populations of the grass Poa alpina, were analyzed using Fourier-transform infrared University of Copenhagen, Denmark (FTIR) spectroscopy, Raman spectroscopy, surface enhanced Raman scattering (SERS), Anna De Juan, and matrix assisted laser desorption/ionization mass spectrometry (MALDI-TOF MS). The University of Barcelona, Spain variation in the sample set can be described in a hierarchical framework comprising three *Correspondence: populations of the same grass species and four different growth conditions of the parent Janina Kneipp [email protected] plants for each of the populations. -

Comparison Between FTIR and Boehm Titration for Activated Carbon Functional Group Quantification
Comparison Between FTIR and Boehm Titration for Activated Carbon Functional Group Quantification Chad Spreadbury, Regina Rodriguez, and David Mazyck College of Engineering, University of Florida Activated carbon (AC) is a proven effective adsorbent of contaminants in the air and water phases. This effectiveness is due to the large surface area of AC which hosts functional groups. Particularly, oxygen functional groups (e.g. carboxyls, lactones, phenols, carbonyls) have been noted as important in mercury removal. Hence, determining the quantities of these groups is crucial when AC is used for this purpose. Currently, Boehm titration is the most common method for determining the number of these functional groups. However, this test method is susceptible to high user error and require a lengthy procedure and run time. On the other hand, Fourier transform infrared (FTIR) spectroscopy excels at analyzing the functionality of an AC surface. This study investigates whether or not a correlation between Boehm titration and FTIR can be determined using quantitative and qualitative assessment. The results indicate that while using the current methodology for Boehm titrations and FTIR analysis, there is no clear correlation. However, this study does not rule out that a correlation may indeed exist between these test methods that can be elucidated with improved methodology that accounts for the unique physical and chemical characteristics of carbon. INTRODUCTION mercury (Hg0) to create oxidized mercury (Hg2+), which then undergoes chemisorption with the AC surface. ctivated carbon is used globally as an effective Boehm titrations and Fourier transform infrared (FTIR) adsorbent for removing contaminants like mercury spectroscopy are two common testing procedures for A(Hg) from air and water phases. -

Venus Elemental and Mineralogical Camera (Vemcam)
EPSC Abstracts Vol. 13, EPSC-DPS2019-827-1, 2019 EPSC-DPS Joint Meeting 2019 c Author(s) 2019. CC Attribution 4.0 license. Venus Elemental and Mineralogical Camera (VEMCam) Samuel M. Clegg (1), Brett S. Okhuysen (1), David S. DeCroix (1), Raymond T. Newell (1), Roger C. Wiens (1), Shiv K. Sharma (2), Sylvestre Maurice (3), Ronald K. Martinez (1), Adriana Reyes-Newell (1), and Melinda D. Dyar (4), (1) Los Alamos National Laboratory, Los Alamos, NM, [email protected], (2) Hawaii Inst. of Geophysics and Planetology, Univ. of Hawaii, Honolulu, USA, (3) L'Institut de Recherche en Astrophysique et Planétologie, Toulouse France, (4) Planetary Science Inst., Tucson, AZ, USA Abstract The Venus Elemental and Mineralogical Camera The Venus Elemental and Mineralogical Camera (VEMCam) is an integrated remote LIBS and Raman (VEMCam) can make thousands of measurements instrument concept designed to operate from within within the first two hours on the surface, providing the safety of the lander. The extreme Venus surface an unprecedented description of the Venus surface. conditions requires rapid analyses and VEMCam can VEMCam is based on the ChemCam instrument on collect over 1000 chemical and mineralogical spectra the Mars Science Laboratory rover and the within the first hour. Here, we discuss the VEMCam SuperCam instrument selected for the Mars 2020 prototype calibration and analysis in which samples rover. VEMCam includes an integrated Raman and are placed in a 2 m long chamber capable of Laser-Induced Breakdown Spectroscopy (LIBS) simulating the Venus surface atmosphere. instrument capable of probing many disparate locations around the lander. VEMCam also includes 1. -

Application of High Performance Liquid Chromatography (HPLC) and Fourier Transform Infrared (FTIR) Spectroscopy to Swiss-Typ
Application of High Performance Liquid Chromatography (HPLC) and Fourier Transform Infrared (FTIR) Spectroscopy to Swiss-type Cheese Split Defects THESIS Presented in Partial Fulfillment of the Requirements for the Master of Science Degree in the Graduate School of The Ohio State University By Feifei Hu Graduate Program in Food Science and Technology The Ohio State University 2012 Master's Examination Committee: Dr. W. James Harper, Advisor Dr. Luis E. Rodriguez-Saona Dr. Michael E. Mangino Copyright Feifei Hu 2012 Abstract Splits, slits and cracks are a continuing problem in the US Swiss Cheese industry and cause downgrading of the cheese, bringing economic loss. The defects generally occur during secondary fermentation in cold storage, which is known to be an issue caused by many factors. Many chemical and physical changes can be indicators to help predicting the defects. Various techniques have been applied to test different compounds in cheese. HPLC is one of the reference methods which have been applied in cheese chemistry for the past decades. Its application involves in the qualification and quantification of organic acids, amino acids and peptides, and sugars in dairy products. However, limited research has been conducted on a rapid and easy testing regimen to investigate split defects in Swiss-type cheese. For over fifty years, researchers have investigated the application of Fourier Transform Infrared (FTIR) Spectroscopy in various food samples, including cheeses and other dairy products because FTIR spectroscopy gives fast, simple and comprehensive detection of chemical compounds. There is great potential for applying FTIR spectroscopy in developing a rapid test method to detect and predict the occurrence of split defects. -

Crystal Structure, Raman Spectroscopy and Dielectric Properties of New Semiorganic Crystals Based on 2-Methylbenzimidazole
crystals Article Crystal Structure, Raman Spectroscopy and Dielectric Properties of New Semiorganic Crystals Based on 2-Methylbenzimidazole E. V. Balashova 1,* , F. B. Svinarev 1, A. A. Zolotarev 2, A. A. Levin 1, P. N. Brunkov 1, V. Yu. Davydov 1 , A. N. Smirnov 1 , A. V. Redkov 3, G. A. Pankova 4 and B. B. Krichevtsov 1 1 Ioffe Institute, 26 Politekhnicheskaya, St Petersburg 194021, Russian Federation; [email protected]ffe.ru (F.B.S.); [email protected]ffe.ru (A.A.L.); [email protected]ffe.ru (P.N.B.); [email protected]ffe.ru (V.Y.D.); [email protected]ffe.ru (A.N.S.); [email protected]ffe.ru (B.B.K.) 2 Institute of Earth Sciences, Saint-Petersburg State University, 7/9 Universitetskaya Nab., St Petersburg 199034, Russian Federation; [email protected] 3 Institute of Problems of Mechanical Engineering, 61 Bolshoy pr. V.O., St Petersburg 199004, Russian Federation; [email protected] 4 Institute of macromolecular compounds, 31 Bolshoy pr. V.O., St Petersburg 199004, Russian Federation; [email protected] * Correspondence: [email protected]ffe.ru Received: 27 September 2019; Accepted: 29 October 2019; Published: 31 October 2019 Abstract: New single crystals, based on 2-methylbenzimidazole (MBI), of MBI-phosphite (C16H24N4O7P2), MBI-phosphate-1 (C16H24N4O9P2), and MBI-phosphate-2 (C8H16N2O9P2) were obtained by slow evaporation method from a mixture of alcohol solution of MBI crystals and water solution of phosphorous or phosphoric acids. Crystal structures and chemical compositions were determined by single crystal X-ray diffraction (XRD) analysis and confirmed by XRD of powders and elemental analysis. -

Article Intends to Provide a for the Necessary Virtual Electronic Brief Overview of the Differences and Transition
ADVANCES IN RAMAN TECHNIQUES Laser requirements and advances for Raman techniques Andreas Isemann Laser Quantum GmbH, 78467 Konstanz, Germany INTRODUCTION 473 nm and 1064 nm, a narrow Raman scattering as a probe of bandwidth output of few tens of GHz vibrational transitions has made or below 1 MHz if needed within the leaps and bounds since its discovery, linewidth of vibrational transitions and various schemes based on this for high resolution, low noise (less phenomenon have been developed than 0.02%) and excellent beam with great success. quality (fundamental transversal Applications range from basic electromagnetic mode TEM00) scientific research, to medical and provides optimised performance industrial instrumentation. Some for the resolution of the Raman schemes utilise linear Raman measurement needed. scattering, whilst others take advantage The wavelength is chosen based of high peak-power fields to probe on the sample under investigation, nonlinear Raman responses. with 532 nm being commonly used This article intends to provide a for the necessary virtual electronic brief overview of the differences and transition. In the following section, benefits, together with the laser source four examples from different areas of requirements and the advancements Raman applications show the diverse in techniques enabled by recent applications of linear Raman and what developments in lasers. advances have been achieved. An example of studying a real-world LINEAR RAMAN application, the successful control of Figure 1 An example of the RR microfluidic device counting of The advent of the laser in providing a food quality using Raman spectroscopy photosynthetic microorganisms. As the cells of the model strain high-intensity coherent light source and multivariate analysis, is described Synechocystis sp. -
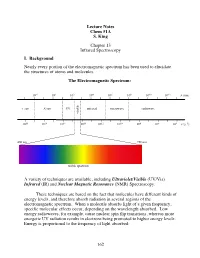
162 Lecture Notes Chem 51A S. King Chapter 13 Infrared Spectroscopy I
Lecture Notes Chem 51A S. King Chapter 13 Infrared Spectroscopy I. Background Nearly every portion of the electromagnetic spectrum has been used to elucidate the structures of atoms and molecules. The Electromagnetic Spectrum: 10!2 100 102 104 106 108 1010 1012 $ (nm) "!ray X-ray UV infrared microwave radiowave visible 1020 1018 1016 1014 1012 1010 108 106 104 # (s!1) 400 nm 750 nm ! visible spectrum A variety of techniques are available, including Ultraviolet/Visible (UV/Vis) Infrared (IR) and Nuclear Magnetic Resonance (NMR) Spectroscopy. These techniques are based on the fact that molecules have different kinds of energy levels, and therefore absorb radiation in several regions of the electromagnetic spectrum. When a molecule absorbs light of a given frequency, specific molecular effects occur, depending on the wavelength absorbed. Low energy radiowaves, for example, cause nuclear spin flip transitions, whereas more energetic UV radiation results in electrons being promoted to higher energy levels. Energy is proportional to the frequency of light absorbed: 162 Molecular effects associated with different regions of the EM spectrum: Wavelength (!) Energy/mole Molecular effects 10"10 meter gamma rays 106 kcal 10"8 meter X-rays 104 kcal ionization vaccum UV 102 kcal near UV electronic transitions 10"6 meter visible 10 kcal infrared 1 kcal molecular vibrations 10"4 meter (IR) 10"2 kcal 10"2 meter microwave rotational motion 10"4 kcal 0 10 meter radio 10"6 kcal nuclear spin transitions 2 10 meter II. IR Spectroscopy IR radiation causes groups of atoms to vibrate with respect to the bonds that connect them.