The Reptile Gut Microbiome: Its Role in Host Evolution and Community Assembly
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
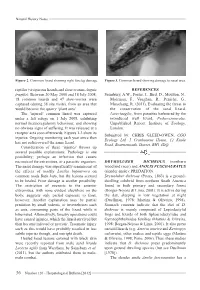
Reptiles (Viviparous Lizards and Slow-Worms Anguis REFERENCES Fragilis)
Natural History Notes Figure 2. Common lizard showing right foreleg damage. Figure 3. Common lizard showing damage to nasal area. reptiles (viviparous lizards and slow-worms Anguis REFERENCES fragilis). Between 20 May 2008 and 18 July 2008, Sainsbury, A.W., Foster, J., Bird, D., Moulton, N., 18 common lizards and 47 slow-worms were Molenaar, F., Vaughan, R., Peniche, G., captured (during 38 site visits), from an area that Marschang, R. (2011). Evaluating the threat to would become the quarry ‘plant area’. the conservation of the sand lizard, The ‘injured’ common lizard was captured Lacertaagilis, from parasites harboured by the under a felt refuge on 1 July 2008, exhibiting introduced wall lizard, Podarcismuralis. normal thermoregulatory behaviour, and showing Unpublished Report. Institute of Zoology, no obvious signs of suffering. It was released at a London. receptor area soon afterwards. Figures 1-3 show its Submitted by: CHRIS GLEED-OWEN, CGO injuries. Ongoing monitoring each year since then Ecology Ltd, 5 Cranbourne House, 12 Knole has not rediscovered the same lizard. Road, Bournemouth, Dorset, BH1 4DQ. Consideration of these ‘injuries’ throws up several possible explanations. Pathology is one possibility; perhaps an infection that causes necrosis of the extremities, or a parasitic organism. DRYMOLUBER DICHROUS (northern The nasal damage was superficially reminiscent of woodland racer) and ANOLIS FUSCOAURATUS the effects of toadfly Lucilia bufonivora on (slender anole): PREDATION. common toads Bufo bufo, but the lesions seemed Drymoluber dichrous (Peters, 1863) is a ground- to be healed. Frost damage is another possibility. dwelling colubrid from northern South America The restriction of necrosis to the anterior found in both primary and secondary forest extremities, with none evident elsewhere on the (Borges-Nojosa & Lima, 2001). -

Phylogenetic Diversity, Habitat Loss and Conservation in South
Diversity and Distributions, (Diversity Distrib.) (2014) 20, 1108–1119 BIODIVERSITY Phylogenetic diversity, habitat loss and RESEARCH conservation in South American pitvipers (Crotalinae: Bothrops and Bothrocophias) Jessica Fenker1, Leonardo G. Tedeschi1, Robert Alexander Pyron2 and Cristiano de C. Nogueira1*,† 1Departamento de Zoologia, Universidade de ABSTRACT Brasılia, 70910-9004 Brasılia, Distrito Aim To analyze impacts of habitat loss on evolutionary diversity and to test Federal, Brazil, 2Department of Biological widely used biodiversity metrics as surrogates for phylogenetic diversity, we Sciences, The George Washington University, 2023 G. St. NW, Washington, DC 20052, study spatial and taxonomic patterns of phylogenetic diversity in a wide-rang- USA ing endemic Neotropical snake lineage. Location South America and the Antilles. Methods We updated distribution maps for 41 taxa, using species distribution A Journal of Conservation Biogeography models and a revised presence-records database. We estimated evolutionary dis- tinctiveness (ED) for each taxon using recent molecular and morphological phylogenies and weighted these values with two measures of extinction risk: percentages of habitat loss and IUCN threat status. We mapped phylogenetic diversity and richness levels and compared phylogenetic distances in pitviper subsets selected via endemism, richness, threat, habitat loss, biome type and the presence in biodiversity hotspots to values obtained in randomized assemblages. Results Evolutionary distinctiveness differed according to the phylogeny used, and conservation assessment ranks varied according to the chosen proxy of extinction risk. Two of the three main areas of high phylogenetic diversity were coincident with areas of high species richness. A third area was identified only by one phylogeny and was not a richness hotspot. Faunal assemblages identified by level of endemism, habitat loss, biome type or the presence in biodiversity hotspots captured phylogenetic diversity levels no better than random assem- blages. -

Other Contributions
Other Contributions NATURE NOTES Amphibia: Caudata Ambystoma ordinarium. Predation by a Black-necked Gartersnake (Thamnophis cyrtopsis). The Michoacán Stream Salamander (Ambystoma ordinarium) is a facultatively paedomorphic ambystomatid species. Paedomorphic adults and larvae are found in montane streams, while metamorphic adults are terrestrial, remaining near natal streams (Ruiz-Martínez et al., 2014). Streams inhabited by this species are immersed in pine, pine-oak, and fir for- ests in the central part of the Trans-Mexican Volcanic Belt (Luna-Vega et al., 2007). All known localities where A. ordinarium has been recorded are situated between the vicinity of Lake Patzcuaro in the north-central portion of the state of Michoacan and Tianguistenco in the western part of the state of México (Ruiz-Martínez et al., 2014). This species is considered Endangered by the IUCN (IUCN, 2015), is protected by the government of Mexico, under the category Pr (special protection) (AmphibiaWeb; accessed 1April 2016), and Wilson et al. (2013) scored it at the upper end of the medium vulnerability level. Data available on the life history and biology of A. ordinarium is restricted to the species description (Taylor, 1940), distribution (Shaffer, 1984; Anderson and Worthington, 1971), diet composition (Alvarado-Díaz et al., 2002), phylogeny (Weisrock et al., 2006) and the effect of habitat quality on diet diversity (Ruiz-Martínez et al., 2014). We did not find predation records on this species in the literature, and in this note we present information on a predation attack on an adult neotenic A. ordinarium by a Thamnophis cyrtopsis. On 13 July 2010 at 1300 h, while conducting an ecological study of A. -

Herpetological Information Service No
Type Descriptions and Type Publications OF HoBART M. Smith, 1933 through June 1999 Ernest A. Liner Houma, Louisiana smithsonian herpetological information service no. 127 2000 SMITHSONIAN HERPETOLOGICAL INFORMATION SERVICE The SHIS series publishes and distributes translations, bibliographies, indices, and similar items judged useful to individuals interested in the biology of amphibians and reptiles, but unlikely to be published in the normal technical journals. Single copies are distributed free to interested individuals. Libraries, herpetological associations, and research laboratories are invited to exchange their publications with the Division of Amphibians and Reptiles. We wish to encourage individuals to share their bibliographies, translations, etc. with other herpetologists through the SHIS series. If you have such items please contact George Zug for instructions on preparation and submission. Contributors receive 50 free copies. Please address all requests for copies and inquiries to George Zug, Division of Amphibians and Reptiles, National Museum of Natural History, Smithsonian Institution, Washington DC 20560 USA. Please include a self-addressed mailing label with requests. Introduction Hobart M. Smith is one of herpetology's most prolific autiiors. As of 30 June 1999, he authored or co-authored 1367 publications covering a range of scholarly and popular papers dealing with such diverse subjects as taxonomy, life history, geographical distribution, checklists, nomenclatural problems, bibliographies, herpetological coins, anatomy, comparative anatomy textbooks, pet books, book reviews, abstracts, encyclopedia entries, prefaces and forwords as well as updating volumes being repnnted. The checklists of the herpetofauna of Mexico authored with Dr. Edward H. Taylor are legendary as is the Synopsis of the Herpetofalhva of Mexico coauthored with his late wife, Rozella B. -

CAT Vertebradosgt CDC CECON USAC 2019
Catálogo de Autoridades Taxonómicas de vertebrados de Guatemala CDC-CECON-USAC 2019 Centro de Datos para la Conservación (CDC) Centro de Estudios Conservacionistas (Cecon) Facultad de Ciencias Químicas y Farmacia Universidad de San Carlos de Guatemala Este documento fue elaborado por el Centro de Datos para la Conservación (CDC) del Centro de Estudios Conservacionistas (Cecon) de la Facultad de Ciencias Químicas y Farmacia de la Universidad de San Carlos de Guatemala. Guatemala, 2019 Textos y edición: Manolo J. García. Zoólogo CDC Primera edición, 2019 Centro de Estudios Conservacionistas (Cecon) de la Facultad de Ciencias Químicas y Farmacia de la Universidad de San Carlos de Guatemala ISBN: 978-9929-570-19-1 Cita sugerida: Centro de Estudios Conservacionistas [Cecon]. (2019). Catálogo de autoridades taxonómicas de vertebrados de Guatemala (Documento técnico). Guatemala: Centro de Datos para la Conservación [CDC], Centro de Estudios Conservacionistas [Cecon], Facultad de Ciencias Químicas y Farmacia, Universidad de San Carlos de Guatemala [Usac]. Índice 1. Presentación ............................................................................................ 4 2. Directrices generales para uso del CAT .............................................. 5 2.1 El grupo objetivo ..................................................................... 5 2.2 Categorías taxonómicas ......................................................... 5 2.3 Nombre de autoridades .......................................................... 5 2.4 Estatus taxonómico -

Xenosaurus Tzacualtipantecus. the Zacualtipán Knob-Scaled Lizard Is Endemic to the Sierra Madre Oriental of Eastern Mexico
Xenosaurus tzacualtipantecus. The Zacualtipán knob-scaled lizard is endemic to the Sierra Madre Oriental of eastern Mexico. This medium-large lizard (female holotype measures 188 mm in total length) is known only from the vicinity of the type locality in eastern Hidalgo, at an elevation of 1,900 m in pine-oak forest, and a nearby locality at 2,000 m in northern Veracruz (Woolrich- Piña and Smith 2012). Xenosaurus tzacualtipantecus is thought to belong to the northern clade of the genus, which also contains X. newmanorum and X. platyceps (Bhullar 2011). As with its congeners, X. tzacualtipantecus is an inhabitant of crevices in limestone rocks. This species consumes beetles and lepidopteran larvae and gives birth to living young. The habitat of this lizard in the vicinity of the type locality is being deforested, and people in nearby towns have created an open garbage dump in this area. We determined its EVS as 17, in the middle of the high vulnerability category (see text for explanation), and its status by the IUCN and SEMAR- NAT presently are undetermined. This newly described endemic species is one of nine known species in the monogeneric family Xenosauridae, which is endemic to northern Mesoamerica (Mexico from Tamaulipas to Chiapas and into the montane portions of Alta Verapaz, Guatemala). All but one of these nine species is endemic to Mexico. Photo by Christian Berriozabal-Islas. amphibian-reptile-conservation.org 01 June 2013 | Volume 7 | Number 1 | e61 Copyright: © 2013 Wilson et al. This is an open-access article distributed under the terms of the Creative Com- mons Attribution–NonCommercial–NoDerivs 3.0 Unported License, which permits unrestricted use for non-com- Amphibian & Reptile Conservation 7(1): 1–47. -

Fossil Amphibians and Reptiles from the Neogene Locality of Maramena (Greece), the Most Diverse European Herpetofauna at the Miocene/Pliocene Transition Boundary
Palaeontologia Electronica palaeo-electronica.org Fossil amphibians and reptiles from the Neogene locality of Maramena (Greece), the most diverse European herpetofauna at the Miocene/Pliocene transition boundary Georgios L. Georgalis, Andrea Villa, Martin Ivanov, Davit Vasilyan, and Massimo Delfino ABSTRACT We herein describe the fossil amphibians and reptiles from the Neogene (latest Miocene or earliest Pliocene; MN 13/14) locality of Maramena, in northern Greece. The herpetofauna is shown to be extremely diverse, comprising at least 30 different taxa. Amphibians include at least six urodelan (Cryptobranchidae indet., Salamandrina sp., Lissotriton sp. [Lissotriton vulgaris group], Lissotriton sp., Ommatotriton sp., and Sala- mandra sp.), and three anuran taxa (Latonia sp., Hyla sp., and Pelophylax sp.). Rep- tiles are much more speciose, being represented by two turtle (the geoemydid Mauremys aristotelica and a probable indeterminate testudinid), at least nine lizard (Agaminae indet., Lacertidae indet., ?Lacertidae indet., aff. Palaeocordylus sp., ?Scin- cidae indet., Anguis sp., five morphotypes of Ophisaurus, Pseudopus sp., and at least one species of Varanus), and 10 snake taxa (Scolecophidia indet., Periergophis micros gen. et sp. nov., Paraxenophis spanios gen. et sp. nov., Hierophis cf. hungaricus, another distinct “colubrine” morphotype, Natrix aff. rudabanyaensis, and another dis- tinct species of Natrix, Naja sp., cf. Micrurus sp., and a member of the “Oriental Vipers” complex). The autapomorphic features and bizarre vertebral morphology of Perier- gophis micros gen. et sp. nov. and Paraxenophis spanios gen. et sp. nov. render them readily distinguishable among fossil and extant snakes. Cryptobranchids, several of the amphibian genera, scincids, Anguis, Pseudopus, and Micrurus represent totally new fossil occurrences, not only for the Greek area, but for the whole southeastern Europe. -

The Sclerotic Ring: Evolutionary Trends in Squamates
The sclerotic ring: Evolutionary trends in squamates by Jade Atkins A Thesis Submitted to Saint Mary’s University, Halifax, Nova Scotia in Partial Fulfillment of the Requirements for the Degree of Master of Science in Applied Science July, 2014, Halifax Nova Scotia © Jade Atkins, 2014 Approved: Dr. Tamara Franz-Odendaal Supervisor Approved: Dr. Matthew Vickaryous External Examiner Approved: Dr. Tim Fedak Supervisory Committee Member Approved: Dr. Ron Russell Supervisory Committee Member Submitted: July 30, 2014 Dedication This thesis is dedicated to my family, friends, and mentors who helped me get to where I am today. Thank you. ! ii Table of Contents Title page ........................................................................................................................ i Dedication ...................................................................................................................... ii List of figures ................................................................................................................. v List of tables ................................................................................................................ vii Abstract .......................................................................................................................... x List of abbreviations and definitions ............................................................................ xi Acknowledgements .................................................................................................... -

Aberrant Colourations in Wild Snakes: Case Study in Neotropical Taxa and a Review of Terminology
SALAMANDRA 57(1): 124–138 Claudio Borteiro et al. SALAMANDRA 15 February 2021 ISSN 0036–3375 German Journal of Herpetology Aberrant colourations in wild snakes: case study in Neotropical taxa and a review of terminology Claudio Borteiro1, Arthur Diesel Abegg2,3, Fabrício Hirouki Oda4, Darío Cardozo5, Francisco Kolenc1, Ignacio Etchandy6, Irasema Bisaiz6, Carlos Prigioni1 & Diego Baldo5 1) Sección Herpetología, Museo Nacional de Historia Natural, Miguelete 1825, Montevideo 11800, Uruguay 2) Instituto Butantan, Laboratório Especial de Coleções Zoológicas, Avenida Vital Brasil, 1500, Butantã, CEP 05503-900 São Paulo, SP, Brazil 3) Universidade de São Paulo, Instituto de Biociências, Departamento de Zoologia, Programa de Pós-Graduação em Zoologia, Travessa 14, Rua do Matão, 321, Cidade Universitária, 05508-090, São Paulo, SP, Brazil 4) Universidade Regional do Cariri, Departamento de Química Biológica, Programa de Pós-graduação em Bioprospecção Molecular, Rua Coronel Antônio Luiz 1161, Pimenta, Crato, Ceará 63105-000, CE, Brazil 5) Laboratorio de Genética Evolutiva, Instituto de Biología Subtropical (CONICET-UNaM), Facultad de Ciencias Exactas Químicas y Naturales, Universidad Nacional de Misiones, Felix de Azara 1552, CP 3300, Posadas, Misiones, Argentina 6) Alternatus Uruguay, Ruta 37, km 1.4, Piriápolis, Uruguay Corresponding author: Claudio Borteiro, e-mail: [email protected] Manuscript received: 2 April 2020 Accepted: 18 August 2020 by Arne Schulze Abstract. The criteria used by previous authors to define colour aberrancies of snakes, particularly albinism, are varied and terms have widely been used ambiguously. The aim of this work was to review genetically based aberrant colour morphs of wild Neotropical snakes and associated terminology. We compiled a total of 115 cases of conspicuous defective expressions of pigmentations in snakes, including melanin (black/brown colour), xanthins (yellow), and erythrins (red), which in- volved 47 species of Aniliidae, Boidae, Colubridae, Elapidae, Leptotyphlopidae, Typhlopidae, and Viperidae. -

Snakes: Cultural Beliefs and Practices Related to Snakebites in a Brazilian Rural Settlement Dídac S Fita1, Eraldo M Costa Neto2*, Alexandre Schiavetti3
Fita et al. Journal of Ethnobiology and Ethnomedicine 2010, 6:13 http://www.ethnobiomed.com/content/6/1/13 JOURNAL OF ETHNOBIOLOGY AND ETHNOMEDICINE RESEARCH Open Access ’Offensive’ snakes: cultural beliefs and practices related to snakebites in a Brazilian rural settlement Dídac S Fita1, Eraldo M Costa Neto2*, Alexandre Schiavetti3 Abstract This paper records the meaning of the term ‘offense’ and the folk knowledge related to local beliefs and practices of folk medicine that prevent and treat snake bites, as well as the implications for the conservation of snakes in the county of Pedra Branca, Bahia State, Brazil. The data was recorded from September to November 2006 by means of open-ended interviews performed with 74 individuals of both genders, whose ages ranged from 4 to 89 years old. The results show that the local terms biting, stinging and pricking are synonymous and used as equivalent to offending. All these terms mean to attack. A total of 23 types of ‘snakes’ were recorded, based on their local names. Four of them are Viperidae, which were considered the most dangerous to humans, besides causing more aversion and fear in the population. In general, local people have strong negative behavior towards snakes, killing them whenever possible. Until the antivenom was present and available, the locals used only charms, prayers and homemade remedies to treat or protect themselves and others from snake bites. Nowadays, people do not pay attention to these things because, basically, the antivenom is now easily obtained at regional hospitals. It is under- stood that the ethnozoological knowledge, customs and popular practices of the Pedra Branca inhabitants result in a valuable cultural resource which should be considered in every discussion regarding public health, sanitation and practices of traditional medicine, as well as in faunistic studies and conservation strategies for local biological diversity. -

Vetagro Sup La Contention Et La Realisation De
VETAGRO SUP CAMPUS VETERINAIRE DE LYON Année 2014 - Thèse n° LA CONTENTION ET LA REALISATION DE PRELEVEMENTS EN VUE D’EXAMENS COMPLEMENTAIRES CHEZ LES REPTILES : REALISATION D’UN CD-ROM INTERACTIF THESE Présentée à l’UNIVERSITE CLAUDE-BERNARD - LYON I (Médecine - Pharmacie) et soutenue publiquement le 19 Décembre 2014 pour obtenir le grade de Docteur Vétérinaire par PETIT Xavier Né le 28 mai 1987 à TROYES VETAGRO SUP CAMPUS VETERINAIRE DE LYON Année 2014 - Thèse n° LA CONTENTION ET LA REALISATION DE PRELEVEMENTS EN VUE D’EXAMENS COMPLEMENTAIRES CHEZ LES REPTILES : REALISATION D’UN CD-ROM INTERACTIF THESE Présentée à l’UNIVERSITE CLAUDE-BERNARD - LYON I (Médecine - Pharmacie) et soutenue publiquement le 19 Décembre 2014 pour obtenir le grade de Docteur Vétérinaire par PETIT Xavier Né le 28 mai 1987 à TROYES 2 Liste des Enseignants du Campus Vétérinaire de Lyon Civilité Nom Prénom Unités pédagogiques Grade M. ALOGNINOUWA Théodore Pathologie du bétail Professeur M. ALVES-DE-OLIVEIRA Laurent Gestion des élevages Maître de conférences Mme ARCANGIOLI Marie-Anne Pathologie du bétail Maître de conférences M. ARTOIS Marc Santé Publique et Vétérinaire Professeur M. BARTHELEMY Anthony Anatomie Chirurgie (ACSAI) Maître de conférences Mme BECKER Claire Pathologie du bétail MaîtreContractuel de conférences M. BELLI Patrick Pathologie morphologique et clinique des animaux de Maître de conférences Mme BENAMOU-SMITH Agnès Equinecompagnie MaîtreContractuel de conférences M. BENOIT Etienne Biologie fonctionnelle Professeur M. BERNY Philippe Biologie fonctionnelle Professeur Mme BERTHELET Marie-Anne Anatomie Chirurgie (ACSAI) Maître de conférences Mme BONNET-GARIN Jeanne-Marie Biologie fonctionnelle Professeur Mme BOULOCHER Caroline Anatomie Chirurgie (ACSAI) Maître de conférences M. -

FIRST CONFIRMED RECORD of the VENOMOUS CORAL SNAKE Micrurus Ibiboboca (SERPENTES, ELAPIDAE) in MINAS GERAIS, BRAZIL
Oecologia Australis 25(1):179-183, 2021 https://doi.org/10.4257/oeco.2021.2501.18 FIRST CONFIRMED RECORD OF THE VENOMOUS CORAL SNAKE Micrurus ibiboboca (SERPENTES, ELAPIDAE) IN MINAS GERAIS, BRAZIL Maria Clara do Nascimento1*, Alexander Zaidan de Souza2, Anderson M. Oliveira3 & Henrique C. Costa4 1 Universidade Federal de Minas Gerais, Instituto de Ciências Biológicas, Departamento de Zoologia. Avenida Antônio Carlos, 6627, CEP: 31270-901, Belo Horizonte, Minas Gerais, Brazil. 2 Fundação Biodiversitas, Avenida Celso Porfírio Machado, 1813, Belvedere, CEP: 30320-400. Belo Horizonte, Minas Gerais, Brazil. 3 Universidade Federal de Viçosa, Departamento de Biologia Animal, Avenida Peter Henry Rolfs s/n, CEP: 36570-900. Viçosa, Minas Gerais, Brazil. 4 Universidade Federal de Juiz de Fora, Departamento de Zoologia. CEP: 36036-900. Juiz de Fora, Minas Gerais, Brazil. E-mail: [email protected] (*corresponding author); [email protected]; anderson.auh@gmail. com; [email protected] Abstract: We report for the first time the occurrence of the venomous snake species Micrurus ibiboboca in the state of Minas Gerais, Brazil, based on a specimen collected in an Atlantic Forest area. This is the sixth record of a venomous coral snake in Minas Gerais and increases to 160 the number of known snake species in the state. Keywords: Atlantic Forest; coral snake; distribution; Squamata; Wallacean shortfall Micrurus ibiboboca (Merrem, 1820) is a coral snake Micrurus ibiboboca is a triadal coral snake (7– species described based on a specimen collected 13 body triads), distinguished from its congeners in the early 19th century by the Prince Maximilian by the following characters: cloacal plate divided; of Wied, in the Atlantic Forest at the mouth of the a black nuchal ring is the anteriormost ring of Belmonte river (now Jequitinhonha river), state the first complete triad; white rings equal to or of Bahia (Campbell & Lamar 2004).