Aligator: a Computational Tool for Optimizing Total Chemical Synthesis of Large Proteins Q ⇑ Michael T
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Peptide Synthesis: Chemical Or Enzymatic
Electronic Journal of Biotechnology ISSN: 0717-3458 Vol.10 No.2, Issue of April 15, 2007 © 2007 by Pontificia Universidad Católica de Valparaíso -- Chile Received June 6, 2006 / Accepted November 28, 2006 DOI: 10.2225/vol10-issue2-fulltext-13 REVIEW ARTICLE Peptide synthesis: chemical or enzymatic Fanny Guzmán Instituto de Biología Pontificia Universidad Católica de Valparaíso Avenida Brasil 2950 Valparaíso, Chile Fax: 56 32 212746 E-mail: [email protected] Sonia Barberis Facultad de Química, Bioquímica y Farmacia Universidad Nacional de San Luis Ejército de los Andes 950 (5700) San Luis, Argentina E-mail: [email protected] Andrés Illanes* Escuela de Ingeniería Bioquímica Pontificia Universidad Católica de Valparaíso Avenida Brasil 2147 Fax: 56 32 2273803 E-mail: [email protected] Financial support: This work was done within the framework of Project CYTED IV.22 Industrial Application of Proteolytic Enzymes from Higher Plants. Keywords: enzymatic synthesis, peptides, proteases, solid-phase synthesis. Abbreviations: CD: circular dichroism CLEC: cross linked enzyme crystals DDC: double dimer constructs ESI: electrospray ionization HOBT: hydroxybenzotriazole HPLC: high performance liquid hromatography KCS: kinetically controlled synthesis MALDI: matrix-assisted laser desorption ionization MAP: multiple antigen peptide system MS: mass spectrometry NMR: nuclear magnetic resonance SPS: solution phase synthesis SPPS: solid-phase peptide synthesis t-Boc: tert-butoxycarbonyl TCS: thermodynamically controlled synthesis TFA: trifluoroacetic acid Peptides are molecules of paramount importance in the medium, biocatalyst and substrate engineering, and fields of health care and nutrition. Several technologies recent advances and challenges in the field are analyzed. for their production are now available, among which Even though chemical synthesis is the most mature chemical and enzymatic synthesis are especially technology for peptide synthesis, lack of specificity and relevant. -

M.Sc. Organic Chemistry
ST. JOSEPH’S COLLEGE (AUTONOMOUS) BENGALURU-27 DEPARTMENT OF CHEMISTRY SYLLABUS FOR POSTGRADUATE COURSE IN ORGANIC CHEMISTRY 2019-21 Re-accredited with ‘A++’ GRADE and 3.79/4 CGPA by NAAC Recognised as “College of Excellence” by UGC The Postgraduate programme in chemistry is designed to give students a good foundation in Chemistry and develop in them problem solving and experimental skills so that they are well prepared for further studies in specialized areas of Chemistry or for employment in academic institutions and in industry. Mission statement: To promote among our learners the skills of thinking, experimentation and application of the knowledge gained. To promote concern for environment and to develop appreciation for green Chemistry. To prepare our students for life in the larger community. Benchmark Statements for the Course: To instil in students a sense of enthusiasm for chemistry, an appreciation of its application in different contexts, and to involve them in intellectually stimulating and satisfying experience of learning and studying. To provide students with a broad and balanced foundation of chemical knowledge and practical skills. Teaching-Learning: Although the lecture method is extensively used, the students are also encouraged to do self- study through other activities like assignments, seminars, quiz, viva-voce etc. Co-curricular Activities: The Chemical Society for postgraduate (P.G.) students provides them with a platform to interact with students of other institutions and also with eminent scientists from -

Polyphenylene Dendrimers
Polyphenylene Dendrimers - Design and Synthesis of Monodisperse Functional Nanoparticles Dissertation zur Erlangung des Grades "Doktor der Wissenschaften" am Fachbereich Chemie und Pharmazie der Johannes Gutenberg-Universität in Mainz vorgelegt von Stefan Bernhardt geb. in Hofheim a. Ts. Mainz 2005 Dekan: Herr Prof. Dr. P. Langguth 1. Berichterstatter: 2. Berichterstatter: Tag der mündlichen Prüfung: 26.06.2006 Die vorliegende Arbeit wurde in der Zeit von März 2002 bis März 2005 am Max-Planck- Institut für Polymerforschung in Mainz unter Anleitung von Herrn Prof. Dr. K. Müllen durchgeführt. Dedicated to my wife If we knew what we were doing it wouldn't be research. Einstein, Albert Index of Abbreviations: AFM atomic force microscopy Anal. analysis calcd. calculated TBAF tetrabutylammoniumfluoride-hexahydrate CD2Cl2 deuterated methylenechloride δ chemical shift / ppm d doublet d8THF deuterated tetrahydrofuran DEE diethylether DMF dimethylformamide DMSO dimethylsulfoxide eq. equivalent EtOH ethanol FD field desorption G1, G2, G3, first-, second-, and third-dendrimer generation GS ground state h hours J coupling constant / Hz LE locally-excited state m multiplett m / g weight in gram MALDI-TOF matrix-assisted laser desorption/ionization-time of flight MCH methylcyclohexane MeOH methanol MS mass spectrometry n refractive index NMR nuclear magnetic resonance PE petroleum ether, low boiling PMI perylene monoimide PPh3 triphenylphosphine ppm parts per million (chemical shift in NMR spectroscopy) RT room temperature s singulet SM single molecule T temperature / °K or °C t triplet TCSPC time-correlated single photon counting TEA triethylamine THF tetrahydrofuran TIPS tri-iso-propylsilyl TPA triphenylamine UV ultraviolett Фflu fluorescence quantum yield τflu decay time of fluorescence kfwd and krev forward and reverse electron transfer rate constants CS charge-separated state Table of contents 1 INTRODUCTION................................................................................................ -

Enantioselective, Convergent Synthesis of the Ineleganolide Core by a Tandem Annulation Cite This: Chem
Chemical Science View Article Online EDGE ARTICLE View Journal | View Issue Enantioselective, convergent synthesis of the ineleganolide core by a tandem annulation Cite this: Chem. Sci.,2017,8,507 cascade† Robert A. Craig, II, Jennifer L. Roizen, Russell C. Smith, Amanda C. Jones, Scott C. Virgil and Brian M. Stoltz* An enantioselective and diastereoselective approach toward the synthesis of the polycyclic norditerpenoid ineleganolide is disclosed. A palladium-catalyzed enantioselective allylic alkylation is employed to stereoselectively construct the requisite chiral tertiary ether and facilitate the synthesis of a 1,3-cis- cyclopentenediol building block. Careful substrate design enabled the convergent assembly of the ineleganolide [6,7,5,5]-tetracyclic scaffold by a diastereoselective cyclopropanation–Cope rearrangement cascade under unusually mild conditions. Computational evaluation of ground state energies of late-stage synthetic intermediates was used to guide synthetic development and aid in the Creative Commons Attribution 3.0 Unported Licence. investigation of the conformational rigidity of these highly constrained and compact polycyclic structures. This work represents the first successful synthesis of the core structure of any member of the furanobutenolide-derived polycyclic norcembranoid diterpene family of natural products. Advanced Received 28th July 2016 synthetic manipulations generated a series of natural product-like compounds that were shown to Accepted 15th August 2016 possess selective secretory antagonism of either interleukin-5 or interleukin-17. This bioactivity stands in DOI: 10.1039/c6sc03347d contrast to the known antileukemic activity of ineleganolide and suggests the norcembranoid natural www.rsc.org/chemicalscience product core may serve as a useful scaffold for the development of diverse therapeutics. This article is licensed under a Introduction this rigid polycyclic scaffold is decorated with a network of nine stereogenic centers, eight of which are contiguous. -

Synthesis and Biosynthesis of Polyketide Natural Products
Syracuse University SURFACE Chemistry - Dissertations College of Arts and Sciences 12-2011 Synthesis and Biosynthesis of Polyketide Natural Products Atahualpa Pinto Syracuse University Follow this and additional works at: https://surface.syr.edu/che_etd Part of the Chemistry Commons Recommended Citation Pinto, Atahualpa, "Synthesis and Biosynthesis of Polyketide Natural Products" (2011). Chemistry - Dissertations. 181. https://surface.syr.edu/che_etd/181 This Dissertation is brought to you for free and open access by the College of Arts and Sciences at SURFACE. It has been accepted for inclusion in Chemistry - Dissertations by an authorized administrator of SURFACE. For more information, please contact [email protected]. Abstract Traditionally separate disciplines of a large and broad chemical spectrum, synthetic organic chemistry and biochemistry have found in the last two decades a fertile common ground in the area pertaining to the biosynthesis of natural products. Both disciplines remain indispensable in providing unique solutions on numerous questions populating the field. Our contributions to this interdisciplinary pursuit have been confined to the biosynthesis of polyketides, a therapeutically and structurally diverse class of natural products, where we employed both synthetic chemistry and biochemical techniques to validate complex metabolic processes. One such example pertained to the uncertainty surrounding the regiochemistry of dehydration and cyclization in the biosynthetic pathway of the marine polyketide spiculoic acid A. The molecule's key intramolecular cyclization was proposed to occur through a linear chain containing an abnormally dehydrated polyene system. We synthesized a putative advanced polyketide intermediate and tested its viability to undergo a mild chemical transformation to spiculoic acid A. In addition, we applied a synthetic and biochemical approach to elucidate the biosynthetic details of thioesterase-catalyzed macrocyclizations in polyketide natural products. -

From Carbon-11-Labeled Amino Acids to Peptides in Positron Emission Tomography: the Synthesis and Clinical Application Aleksandra Pekošak, Ulrike Filp, Alex J
Mol Imaging Biol (2018) DOI: 10.1007/s11307-018-1163-5 * The Author(s), 2018. This article is an open access publication REVIEW ARTICLE From Carbon-11-Labeled Amino Acids to Peptides in Positron Emission Tomography: the Synthesis and Clinical Application Aleksandra Pekošak, Ulrike Filp, Alex J. Poot, Albert D. Windhorst Radionuclide Center, Department of Radiology and Nuclear Medicine, VU University Medical Center, De Boelelaan 1085c, 1081 HV, Amsterdam, The Netherlands Abstract Radiolabeled amino acids, their derivatives and peptides have a broad scope of application and can be used as receptor ligands, as well as enzyme substrates for many different diseases as radiopharmaceutical tracers. Over the past few decades, the application of molecular imaging techniques such as positron emission tomography (PET) has gained considerable importance and significance in diagnosis in today’s advanced health care. Next to that, the availability of cyclotrons and state-of-the-art radiochemistry facilities has progressed the production of imaging agents enabling the preparation of many versatile PET radiotracers. Due to many favorable characteristics of radiolabeled amino acids and peptides, they can be used for tumor staging and monitoring the progress of therapy success, while aromatic amino acids can be employed as PET tracer to study neurological disorders. This review provides a comprehensive overview of radiosynthetic and enzymatic approaches towards carbon-11 amino acids, their analogues and peptides, with focus on stereoselective reactions, and reflects upon their clinical application. Key Words: Carbon-11, Amino acid, Peptide, Radiolabeling, PET imaging Introduction D-glucose ([18F]FDG) as well as a large variety of other PET tracers like receptor ligands or enzyme substrates. -

Convergent Synthesis of Thioether Containing Peptides
molecules Article Convergent Synthesis of Thioether Containing Peptides Spyridon Mourtas 1,2, Christina Katakalou 1, Dimitrios Gatos 1 and Kleomenis Barlos 1,3,* 1 Department of Chemistry, University of Patras, 26510 Rio Patras, Greece; [email protected] (S.M.); [email protected] (C.K.); [email protected] (D.G.) 2 Institute of Chemical Engineering Sciences of the Foundation for Research and Technology Hellas (FORTH/IEC-HT), 26504 Rio Patras, Greece 3 CBL-Patras, Patras Industrial Area, Block 1, 25018 Patras, Greece * Correspondence: [email protected]; Tel.: +30-2610-647600; Fax: +30-2610-647316 Academic Editor: Theodore Tselios Received: 23 December 2019; Accepted: 3 January 2020; Published: 5 January 2020 Abstract: Thioether containing peptides were obtained following three synthetic routes. In route A, halo acids esterified on 2-chlorotrityl(Cltr) resin were reacted with N-fluorenylmethoxycarbonyl (Fmoc) aminothiols. These were either cleaved from the resin to the corresponding (Fmoc-aminothiol) carboxylic acids, which were used as key building blocks in solid phase peptide synthesis (SPPS), or the N-Fmoc group was deprotected and peptide chains were elongated by standard SPPS. The obtained N-Fmoc protected thioether containing peptides were then condensed either in solution, or on solid support, with the appropriate amino components of peptides. In route B, the thioether containing peptides were obtained by the reaction of N-Fmoc aminothiols with bromoacetylated peptides, which were synthesized on Cltr-resin, followed by removal of the N-Fmoc group and subsequent peptide elongation by standard SPPS. In route C, the thioether containing peptides were obtained by the condensation of a haloacylated peptide synthesized on Cltr-resin and a thiol-peptide synthesized either on 4-methoxytrityl(Mmt) or trityl(Trt) resin. -

CV-PSB-August 2020
Curriculum Vitae Phil S. Baran Appointment: Scripps Research Professor, Department of Chemistry 10550 North Torrey Pines Road, BCC-436 La Jolla, California 92037 Telephone: (858) 784-7373 Facsimile: (858) 784-7575 Email: [email protected] Website: www.scripps.edu/chem/baran/ January, 2013 Darlene Shiley Professor of Chemistry April, 2009 Member, Skaggs Institute for Chemical Biology June, 2008 Professor of Chemistry July, 2006 Associate Professor of Chemistry (with Tenure) June, 2003 Assistant Professor of Chemistry Date/Place of Birth: 10 Aug 1977 / Denville, NJ, USA Citizenship: United States Education 2001 – 2003 Postdoctoral Associate Advisor: Professor E.J. Corey Harvard University, Cambridge, Massachusetts 1997 – 2001 Ph.D. Graduate Student in Chemistry Advisor: Professor K.C. Nicolaou The Scripps Research Institute, La Jolla, California 1995 – 1997 B.S. with Honors in Chemistry Advisor: Professor D.I. Schuster New York University, New York, New York 1991 – 1995 Simultaneous high school graduation from Mt. Dora High School and A.A. degree with honors, Lake Sumter Community College, Florida Awards • Inhoffen Medal, 2019 • Manchot Research Professorship, 2017 • Member, The National Academy of Sciences, 2017 • Emanuel Merck Lectureship, 2017 • Blavatnik National Laureate in Chemistry, 2016 • ACS Elias J. Corey Award, 2016 • Member, American Academy of Arts and Sciences, 2015 • College of Arts and Science Alumni Distinguished Service Award, New York University, 2015 • Reagent of the Year Award (EROS), 2015 • Mukaiyama Award, 2014 • -

A Review on Current Industrial Trends for Synthesis of Medicinal Compounds
AAAcccaaadddeeemmmiiiccc SSSccciiieeennnccceeesss International Journal of Pharmacy and Pharmaceutical Sciences ISSN- 0975-1491 Vol 5, Issue 4, 2013 Review Article A REVIEW ON CURRENT INDUSTRIAL TRENDS FOR SYNTHESIS OF MEDICINAL COMPOUNDS SMITA JAIN, I.G. RATHISH, R. SANKARAN* Fresenius Kabi Oncology Ltd., Echelon Institutional Area, Plot No. 11, Sector 32, Gurgaon – 122001, Haryana, India. Email: [email protected] Received: 26 Jul 2013, Revised and Accepted: 18 Aug 2013 ABSTRACT In the pharmaceutical industry, computational methods have gained a major role in the discovery and development of new drugs. Yet, synthesis of the pharmaceutical compounds is among the most crucial steps of drug design along with the life science discoveries complementing it. The review focuses on all the recent developments in the application of computational methods as well as organic synthetic methodology in the field of pharmaceutical research. The most modern synthetic developments of pharmacologically interesting compounds (carbohydrates and nucleotides) as well as important synthetic methods such as combinatorial chemistry, solid-phase reactions, bio-assisted organic synthesis and asymmetric synthesis as applicable to drug discovery and development are critically discussed. Keywords: Lead Discovery, Computational chemistry, Asymmetric synthesis, Biocatalysis, Green synthesis. Drugs play important and a vital role in everybody’s life in today’s 1.1 Computational Drug Design: world. This is what drives to set up new R&Ds in pharmaceutical industry to study and develop knowledge databases on diseases, Against the traditional trial and error based synthetic approach comprising synthesis of drug candidates and analyzing the biological mechanisms treatment and medicines. Undoubtedly, pharmaceutical activity blindly, a more rational, million-saving and high-throughput research is an interdisciplinary area and needs successful integration of approach has developed with the advent of the computers. -
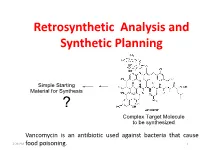
Retrosynthetic Analysis and Synthetic Planning
Retrosynthetic Analysis and Synthetic Planning Vancomycin is an antibiotic used against bacteria that cause 2:24 PM food poisoning. 1 Life’s Perspectives Planning a Journey to the Unknown 2:24 PM 2 Retrosynthetic Analysis Definition Retrosynthetic analysis (retrosynthesis) is a technique for planning a synthesis, especially of complex organic molecules, whereby the complex target molecule (TM) is reduced into a sequence of progressively simpler structures along a pathway which ultimately leads to the identification of a simple or commercially available starting material (SM) from which a chemical synthesis can then be developed. Retrosynthetic analysis is based on known reactions (e.g the Wittig reaction, oxidation, reduction etc). The synthetic plan generated from the retrosynthetic analysis will be the roadmap to guide the synthesis of the target molecule. 2:24 PM 3 Synthetic Planning Definition Synthesis is a construction process that involves converting simple or commercially available molecules into complex molecules using specific reagents associated with known reactions in the retrosynthetic scheme. Syntheses can be grouped into two broad categories: (i) Linear syntheses 2:24 PM (ii)Convergent syntheses 4 Linear Synthesis Definition In linear synthesis, the target molecule is synthesized through a series of linear transformations. Since the overall yield of the synthesis is based on the single longest route to the target molecule, by being long, a linear synthesis suffers a lower overall yield. The linear synthesis is fraught with failure for its lack of flexibility leading to potential large losses in the material already invested in the synthesis at the time of failure. 2:24 PM 5 Convergent Synthesis Definition In convergent synthesis, key fragments of the target molecule are synthesized separately or independently and then brought together at a later stage in the synthesis to make the target molecule. -

Convergent Total Synthesis and Preliminary Biological Investigations
Norrislide: Convergent Total Synthesis and Preliminary Biological Investigations Author: Krista Elizabeth Granger Persistent link: http://hdl.handle.net/2345/731 This work is posted on eScholarship@BC, Boston College University Libraries. Boston College Electronic Thesis or Dissertation, 2009 Copyright is held by the author, with all rights reserved, unless otherwise noted. Boston College The Graduate School of Arts and Sciences Department of Chemistry NORRISOLIDE: CONVERGENT TOTAL SYNTHESIS AND PRELIMINARY BIOLOGICAL INVESTIGATIONS a dissertation by KRISTA ELIZABETH GRANGER submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy August 2009 © copyright by KRISTA ELIZABETH GRANGER 2009 Norrisolide: Convergent Total Synthesis and Preliminary Biological Investigations Krista Elizabeth Granger Thesis Advisor: Professor Marc L. Snapper Abstract • Chapter 1: A review of Shapiro reactions as a coupling strategy in natural product total synthesis. The syntheses of lycoramine, galanthamine, yuehchukene analogues, ovalicin, studies toward the ingenol core, haemanthidine, pretazettine, tazettine, crinamine, Taxol, colombiasin A, elisapterosin B, the AB ring fragment of spongistatin 1 and 8-epipuupewhedione are discussed. Ar O S O nBuLi Li E+ E NH R' R' N R R R' R • Chapter 2: The convergent total synthesis of the marine natural product norrisolide is described. Both subunits, the hydrindane core and the norrisane side chain, are prepared in an asymmetric fashion through kinetic resolution and enantioselective cyclopropanation, respectively. A Shapiro reaction couples the two fragments and a Peterson olefination installs the 1,1-disubstituted olefin. O O MeO OTBS AcO MeO O O O O N Me Me Me MeO O O Me Li O OP H H Me Me Me Me norrisolide H Me Me • Chapter 3: Preliminary experiments to isolate the biological target of norrisolide through reductive alkylation and tritium labeling are investigated. -

Design and Synthesis of Handles for Solid-Phase Peptide Synthesis And
Louisiana State University LSU Digital Commons LSU Doctoral Dissertations Graduate School 2003 Design and synthesis of handles for solid-phase peptide synthesis and convergent peptide synthesis Jose Giraldes Louisiana State University and Agricultural and Mechanical College, [email protected] Follow this and additional works at: https://digitalcommons.lsu.edu/gradschool_dissertations Part of the Chemistry Commons Recommended Citation Giraldes, Jose, "Design and synthesis of handles for solid-phase peptide synthesis and convergent peptide synthesis" (2003). LSU Doctoral Dissertations. 1146. https://digitalcommons.lsu.edu/gradschool_dissertations/1146 This Dissertation is brought to you for free and open access by the Graduate School at LSU Digital Commons. It has been accepted for inclusion in LSU Doctoral Dissertations by an authorized graduate school editor of LSU Digital Commons. For more information, please [email protected]. DESIGN AND SYNTHESIS OF HANDLES FOR SOLID-PHASE PEPTIDE SYNTHESIS AND CONVERGENT PEPTIDE SYNTHESIS A Dissertation Submitted to the Graduate Faculty of the Louisiana State University and Agricultural and Mechanical College in partial fulfillment of the requirements for the degree of Doctor of Philosophy in The Department of Chemistry by José Giraldés B.S., University of Puerto Rico, 1997 May, 2003 To my family ii ACKNOWLEDGMENTS I would like to thank my major professor Dr. Mark McLaughlin for his invaluable guidance and support during my stay at LSU. I am very grateful for the freedom and encouragement he gave me to develop my own ideas. A major amount of thanks must be given to Dr. Frank Zhou for the magic angle spinning NMR, to Martha Juban for help with peptide synthesis and purification, to Dr.