Plasma Methylation Profile RESOURCE GUIDE
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

S41598-021-96252-4.Pdf
www.nature.com/scientificreports OPEN Comprehensive polar metabolomics and lipidomics profling discriminates the transformed from the non‑transformed state in colon tissue and cell lines Caroline Rombouts1,2,3, Margot De Spiegeleer1, Lieven Van Meulebroek1, Lynn Vanhaecke1,4,5* & Winnok H. De Vos3,5* Colorectal cancer (CRC) is the fourth most lethal disease worldwide. Despite an urgent need for therapeutic advance, selective target identifcation in a preclinical phase is hampered by molecular and metabolic variations between cellular models. To foster optimal model selection from a translational perspective, we performed untargeted ultra‑high performance liquid chromatography coupled to high‑resolution mass spectrometry‑based polar metabolomics and lipidomics to non‑ transformed (CCD841‑CON and FHC) and transformed (HCT116, HT29, Caco2, SW480 and SW948) colon cell lines as well as tissue samples from ten colorectal cancer patients. This unveiled metabolic signatures discriminating the transformed from the non‑transformed state. Metabolites involved in glutaminolysis, tryptophan catabolism, pyrimidine, lipid and carnitine synthesis were elevated in transformed cells and cancerous tissue, whereas those involved in the glycerol‑3‑phosphate shuttle, urea cycle and redox reactions were lowered. The degree of glutaminolysis and lipid synthesis was specifc to the colon cancer cell line at hand. Thus, our study exposed pathways that are specifcally associated with the transformation state and revealed diferences between colon cancer cell lines that should be considered when targeting cancer‑associated pathways. Colorectal cancer (CRC) is the second and third most diagnosed cancer in females and males, respectively, and the fourth leading cause of cancer-related mortality worldwide. Te incidence rates are strongly variable throughout the world, whereby developed regions have more CRC patients than less developed countries. -

Methionine Synthase Supports Tumor Tetrahydrofolate Pools
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.05.284521; this version posted September 7, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Methionine synthase supports tumor tetrahydrofolate pools Joshua Z. Wang1,2,#, Jonathan M. Ghergurovich1,3,#, Lifeng Yang1,2, and Joshua D. Rabinowitz1,2,* 1 Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, New Jersey, USA 2 Department of Chemistry, Princeton University, Princeton, New Jersey, USA 3 Department of Molecular Biology, Princeton University, Princeton, New Jersey, USA # These authors contributed equally to this work. *Corresponding author: Joshua Rabinowitz Department of Chemistry and the Lewis-Sigler Institute for Integrative Genomics, Princeton University, Washington Rd, Princeton, NJ 08544, USA Phone: (609) 258-8985; e-mail: [email protected] Conflict of Interest Disclosure: J.D.R. is a paid advisor and stockholder in Kadmon Pharmaceuticals, L.E.A.F. Pharmaceuticals, and Rafael Pharmaceuticals; a paid consultant of Pfizer; a founder, director, and stockholder of Farber Partners and Serien Therapeutics. JDR and JMG are inventors of patents in the area of folate metabolism held by Princeton University. 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.09.05.284521; this version posted September 7, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Mammalian cells require activated folates to generate nucleotides for growth and division. The most abundant circulating folate species is 5-methyl tetrahydrofolate (5-methyl- THF), which is used to synthesize methionine from homocysteine via the cobalamin-dependent enzyme methionine synthase (MTR). -

Defining Sources of Nutrient Limitation for Tumors By
Defining sources of nutrient limitation for tumors by Mark Robert Sullivan B.S. Molecular Biology and Chemistry University of Pittsburgh (2013) Submitted to the Department of Biology in Partial Fulfillment of the Requirements for the Degree of DOCTOR OF PHILOSOPHY at the MASSACHUSETTS INSTITUTE OF TECHNOLOGY June 2019 © 2019 Mark R. Sullivan. All rights reserved. The author hereby grants to MIT permission to reproduce and to distribute publicly paper and electronic copies of this thesis document in whole or in part in any medium now known or hereafter created. Signature of Author.......................................................................................................................... Department of Biology April 30, 2019 Certified by ...................................................................................................................................... Matthew G. Vander Heiden Associate Professor of Biology Thesis Supervisor Accepted by...................................................................................................................................... Amy E. Keating Professor of Biology Co-Director, Biology Graduate Committee 1 2 Defining sources of nutrient limitation for tumors by Mark Robert Sullivan Submitted to the Department of Biology on April 30, 2019 in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy in Biology ABSTRACT Tumor growth requires that cancer cells accumulate sufficient biomass to grow and divide. To accomplish this, tumor cells must acquire -

University Microfilms International 300 N
INFORMATION TO USERS This was produced from a copy of a document sent to us for microfilming. While the most advanced technological means to photograph and reproduce this document have been used, the quality is heavily dependent upon the quality of the material submitted. The following explanation of techniques is provided to help you understand markings or notations which may appear on this reproduction. 1. The sign or "target” for pages apparently lacking from the document photographed is "Missing Page(s)”. If it was possible to obtain the missing page(s) or section, they are spliced into the film along with adjacent pages. This may have necessitated cutting through an image and duplicating adjacent pages to assure you of complete continuity. 2. When an image on the film is obliterated with a round black mark it is an indication that the film inspector noticed either blurred copy because of movement during exposure, or duplicate copy. Unless we meant to delete copyrighted materials that should not have been filmed, you will find a good image of the page in the adjacent frame. If copyrighted materials were deleted you will find a target note listing the pages in the adjacent frame. 3. When a map, drawing or chart, etc., is part of the material being photo graphed the photographer has followed a definite method in “sectioning” the material. It is customary to begin filming at the upper left hand corner of a large sheet and to continue from left to right in equal sections with small overlaps. If necessary, sectioning is continued again—beginning below the first row and continuing on until complete. -

Dysregulated Hepatic Methionine
Virginia Commonwealth University VCU Scholars Compass Internal Medicine Publications Dept. of Internal Medicine 2015 Dysregulated Hepatic Methionine Metabolism Drives Homocysteine Elevation in Diet-Induced Nonalcoholic Fatty Liver Disease Tommy Pacana Virginia Commonwealth University, [email protected] Sophie Cazanave Virginia Commonwealth University Aurora Verdianelli Virginia Commonwealth University See next page for additional authors Follow this and additional works at: http://scholarscompass.vcu.edu/intmed_pubs Part of the Medicine and Health Sciences Commons Copyright: © 2015 Pacana et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited Downloaded from http://scholarscompass.vcu.edu/intmed_pubs/98 This Article is brought to you for free and open access by the Dept. of Internal Medicine at VCU Scholars Compass. It has been accepted for inclusion in Internal Medicine Publications by an authorized administrator of VCU Scholars Compass. For more information, please contact [email protected]. Authors Tommy Pacana, Sophie Cazanave, Aurora Verdianelli, Viashali Patel, Hae-Ki Min, Faridoddin Mirshahi, Eoin Quinlavin, and Arun J. Sanyal This article is available at VCU Scholars Compass: http://scholarscompass.vcu.edu/intmed_pubs/98 RESEARCH ARTICLE Dysregulated Hepatic Methionine Metabolism Drives Homocysteine Elevation in Diet-Induced Nonalcoholic Fatty -
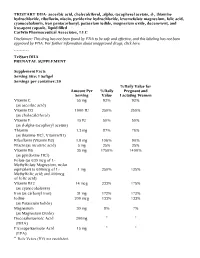
TRISTART DHA- Ascorbic Acid, Cholecalciferol, .Alpha
TRISTART DHA- ascorbic acid, cholecalciferol, .alpha.-tocopherol acetate, d-, thiamine hydrochloride, riboflavin, niacin, pyridoxine hydrochloride, levomefolate magnesium, folic acid, cyanocobalamin, iron pentacarbonyl, potassium iodide, magnesium oxide, doconexent, and icosapent capsule, liquid filled CarWin Pharmaceutical Associates, LLC Disclaimer: This drug has not been found by FDA to be safe and effective, and this labeling has not been approved by FDA. For further information about unapproved drugs, click here. ---------- TriStart DHA PRENATAL SUPPLEMENT Supplement Facts Serving Size: 1 Softgel Servings per container: 30 %Daily Value for Amount Per %Daily Pregnant and Serving Value Lactating Women Vitamin C 55 mg 92% 92% (as ascorbic acid) Vitamin D3 1000 IU 250% 250% (as cholecalciferol) Vitamin E 15 IU 50% 50% (as d-alpha-tocopheryl acetate) Thiamin 1.3 mg 87% 76% (as thiamine HCl, Vitamin B1) Riboflavin (Vitamin B2) 1.8 mg 106% 90% Niacin (as nicotinic acid) 5 mg 25% 25% Vitamin B6 35 mg 1750% 1400% (as pyridoxine HCl) Folate (as 630 mcg of L- Methylfolate Magnesium, molar equivalent to 600mcg of L- 1 mg 250% 125% Methylfolic acid; and 400mcg of folic acid) Vitamin B12 14 mcg 233% 175% (as cyanocobalamin) Iron (as carbonyl iron) 31 mg 172% 172% Iodine 200 mcg 133% 133% (as Potassium Iodide) Magnesium 30 mg 8% 7% (as Magnesium Oxide) Docosahexaenoic Acid 200mg * * (DHA) Eicosapentaenoate Acid 15 mg * * (EPA) * Daily Values (DV) not established. OTHER INGREDIENTS: Gelatin (bovine), Glycerin, Purified Water, Yellow Bees Wax, Caramel Powder, Soy Lecithin, Natural Orange Flavor, Ethyl Vanillin. Contains: Soy and Fish TriStart DHA™ Softgel capsules are dye free, lactose, gluten and sugar free. -

Cysteine Dioxygenase 1 Is a Metabolic Liability for Non-Small Cell Lung Cancer Authors: Yun Pyo Kang1, Laura Torrente1, Min Liu2, John M
bioRxiv preprint doi: https://doi.org/10.1101/459602; this version posted November 1, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Cysteine dioxygenase 1 is a metabolic liability for non-small cell lung cancer Authors: Yun Pyo Kang1, Laura Torrente1, Min Liu2, John M. Asara3,4, Christian C. Dibble5,6 and Gina M. DeNicola1,* Affiliations: 1 Department of Cancer Physiology, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL, USA 2 Proteomics and Metabolomics Core Facility, Moffitt Cancer Center and Research Institute, Tampa, FL, USA 3 Division of Signal Transduction, Beth Israel Deaconess Medical Center, Boston, MA, USA 4 Department of Medicine, Harvard Medical School, Boston, MA, USA 5 Department of Pathology and Cancer Center, Beth Israel Deaconess Medical Center, Boston, MA, USA 6 Department of Pathology, Harvard Medical School, Boston, MA, USA *Correspondence to: [email protected]. Keywords: KEAP1, NRF2, cysteine, CDO1, sulfite Summary NRF2 is emerging as a major regulator of cellular metabolism. However, most studies have been performed in cancer cells, where co-occurring mutations and tumor selective pressures complicate the influence of NRF2 on metabolism. Here we use genetically engineered, non-transformed primary cells to isolate the most immediate effects of NRF2 on cellular metabolism. We find that NRF2 promotes the accumulation of intracellular cysteine and engages the cysteine homeostatic control mechanism mediated by cysteine dioxygenase 1 (CDO1), which catalyzes the irreversible metabolism of cysteine to cysteine sulfinic acid (CSA). Notably, CDO1 is preferentially silenced by promoter methylation in non-small cell lung cancers (NSCLC) harboring mutations in KEAP1, the negative regulator of NRF2. -

RT² Profiler PCR Array (Rotor-Gene® Format) Human Amino Acid Metabolism I
RT² Profiler PCR Array (Rotor-Gene® Format) Human Amino Acid Metabolism I Cat. no. 330231 PAHS-129ZR For pathway expression analysis Format For use with the following real-time cyclers RT² Profiler PCR Array, Rotor-Gene Q, other Rotor-Gene cyclers Format R Description The Human Amino Acid Metabolism I RT² Profiler PCR Array profiles the expression of 84 key genes important in biosynthesis and degradation of functional amino acids. Of the 20 amino acids required for protein synthesis, six of them (arginine, cysteine, glutamine, leucine, proline, and tryptophan), collectively known as the functional amino acids, regulate key metabolic pathways involved in cellular growth, and development, as well as other important biological processes such as immunity and reproduction. For example, leucine activates mTOR signaling and increases protein synthesis, leading to lymphocyte proliferation. Therefore, a lack of leucine can compromise immune function. Metabolic pathways interrelated with the biosynthesis and degradation of these amino acids include vitamin and cofactor biosynthesis (such as SAM or S-Adenosyl Methionine) as well as neurotransmitter metabolism (such as glutamate). This array includes genes for mammalian functional amino acid metabolism as well as genes involved in methionine metabolism, important also for nutrient sensing and sulfur metabolism. Using realtime PCR, you can easily and reliably analyze the expression of a focused panel of genes involved in functional amino acid metabolism with this array. For further details, consult the RT² Profiler PCR Array Handbook. Shipping and storage RT² Profiler PCR Arrays in the Rotor-Gene format are shipped at ambient temperature, on dry ice, or blue ice packs depending on destination and accompanying products. -

Sarcosine and Other Metabolites Along the Choline Oxidation Pathway in Relation to Prostate Cancer—A Large Nested Case–Control Study Within the JANUS Cohort in Norway
IJC International Journal of Cancer Sarcosine and other metabolites along the choline oxidation pathway in relation to prostate cancer—A large nested case–control study within the JANUS cohort in Norway Stefan de Vogel1, Arve Ulvik2, Klaus Meyer2, Per Magne Ueland3,4, Ottar Nyga˚rd5,6, Stein Emil Vollset1,7, Grethe S. Tell1, Jesse F. Gregory III8, Steinar Tretli9 and Tone Bjïrge1,7 1 Department of Global Public Health and Primary Care, University of Bergen, Bergen, Norway 2 BEVITAL AS, Bergen, Norway 3 Section for Pharmacology, Institute of Medicine, University of Bergen, Bergen, Norway 4 Laboratory of Clinical Biochemistry, Haukeland University Hospital, Bergen, Norway 5 Section for Cardiology, Institute of Medicine, University of Bergen, Bergen, Norway 6 Department of Heart Disease, Haukeland University Hospital, Bergen, Norway 7 Norwegian Institute of Public Health, Bergen, Norway 8 Food Science and Human Nutrition Department, University of Florida, Gainesville, FL 9 The Cancer Registry of Norway, Oslo, Norway Methyl group donors and intermediates of one-carbon metabolism affect DNA synthesis and DNA methylation, and may thereby affect prostate carcinogenesis. Choline, the precursor of betaine, and the one-carbon metabolite sarcosine have been associated with increased prostate cancer risk. Within JANUS, a prospective cohort in Norway (n 5 317,000) with baseline serum samples, we conducted a nested case–control study among 3,000 prostate cancer cases and 3,000 controls. Using conditional logistic regression, odds ratios (ORs) and 95% confidence intervals (CIs) for prostate cancer risk were estimated according to quintiles of circulating betaine, dimethylglycine (DMG), sarcosine, glycine and serine. High sarcosine and glycine concentrations were associated with reduced prostate cancer risk of borderline significance (sarcosine: highest vs. -

The Different Catalytic Roles of the Metal Binding Ligands in Human 4- Hydroxyphenylpyruvate Dioxygenase
Citation for published version: Huang, C-W, Liu, H-C, Shen, C-P, Chen, Y-T, Lee, S-J, Lloyd, MD & Lee, H-J 2016, 'The different catalytic roles of the metal- binding ligands in human 4-hydroxyphenylpyruvate dioxygenase', Biochemical Journal, vol. 473, no. 9, pp. 1179-1189. https://doi.org/10.1042/BCJ20160146 DOI: 10.1042/BCJ20160146 Publication date: 2016 Document Version Peer reviewed version Link to publication Publisher Rights Unspecified University of Bath Alternative formats If you require this document in an alternative format, please contact: [email protected] General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Download date: 28. Sep. 2021 BIOCHEMICAL JOURNAL ACCEPTED MANUSCRIPT The different catalytic roles of the metal binding ligands in human 4- hydroxyphenylpyruvate dioxygenase Chih‐Wei Huang, Hsiu‐Chen Liu, Chia‐Pei Shen, Yi‐Tong Chen, Sung‐Jai Lee, Matthew D. Lloyd, and Hwei‐Jen Lee 4‐Hydroxylphenylpyruvate dioxygenase (HPPD) is a non‐haem iron(II)‐dependent oxygenase that catalyzes the conversion of 4‐hydroxylphenylpyruvate (HPP) to homogentisate (HG). In the active site, a strictly conserved 2‐His‐1‐Glu facial triad coordinates the iron ready for catalysis. Substitution of these residues resulted in about a 10‐fold decrease in the metal binding affinity, as measured by isothermal titration calorimetry, and a large reduction in enzyme catalytic efficiencies. -

1 Abietic Acid R Abrasive Silica for Polishing DR Acenaphthene M (LC
1 abietic acid R abrasive silica for polishing DR acenaphthene M (LC) acenaphthene quinone R acenaphthylene R acetal (see 1,1-diethoxyethane) acetaldehyde M (FC) acetaldehyde-d (CH3CDO) R acetaldehyde dimethyl acetal CH acetaldoxime R acetamide M (LC) acetamidinium chloride R acetamidoacrylic acid 2- NB acetamidobenzaldehyde p- R acetamidobenzenesulfonyl chloride 4- R acetamidodeoxythioglucopyranose triacetate 2- -2- -1- -β-D- 3,4,6- AB acetamidomethylthiazole 2- -4- PB acetanilide M (LC) acetazolamide R acetdimethylamide see dimethylacetamide, N,N- acethydrazide R acetic acid M (solv) acetic anhydride M (FC) acetmethylamide see methylacetamide, N- acetoacetamide R acetoacetanilide R acetoacetic acid, lithium salt R acetobromoglucose -α-D- NB acetohydroxamic acid R acetoin R acetol (hydroxyacetone) R acetonaphthalide (α)R acetone M (solv) acetone ,A.R. M (solv) acetone-d6 RM acetone cyanohydrin R acetonedicarboxylic acid ,dimethyl ester R acetonedicarboxylic acid -1,3- R acetone dimethyl acetal see dimethoxypropane 2,2- acetonitrile M (solv) acetonitrile-d3 RM acetonylacetone see hexanedione 2,5- acetonylbenzylhydroxycoumarin (3-(α- -4- R acetophenone M (LC) acetophenone oxime R acetophenone trimethylsilyl enol ether see phenyltrimethylsilyl... acetoxyacetone (oxopropyl acetate 2-) R acetoxybenzoic acid 4- DS acetoxynaphthoic acid 6- -2- R 2 acetylacetaldehyde dimethylacetal R acetylacetone (pentanedione -2,4-) M (C) acetylbenzonitrile p- R acetylbiphenyl 4- see phenylacetophenone, p- acetyl bromide M (FC) acetylbromothiophene 2- -5- -

Separation and Quantitation of Water Soluble Cellular Metabolites by Hydrophilic Interaction Chromatography-Tandem Mass Spectrometry Sunil U
Journal of Chromatography A, 1125 (2006) 76–88 Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry Sunil U. Bajad a, Wenyun Lu a, Elizabeth H. Kimball a, Jie Yuan a, Celeste Peterson b, Joshua D. Rabinowitz a,∗ a Lewis-Sigler Institute for Integrative Genomics and Department of Chemistry, Princeton University, Princeton, NJ 08544, USA b Department of Molecular Biology, Princeton University, Princeton, NJ 08544, USA Received 13 January 2006; received in revised form 8 May 2006; accepted 10 May 2006 Available online 6 June 2006 Abstract A key unmet need in metabolomics is the ability to efficiently quantify a large number of known cellular metabolites. Here we present a liquid chromatography (LC)–electrospray ionization tandem mass spectrometry (ESI-MS/MS) method for reliable measurement of 141 metabolites, including components of central carbon, amino acid, and nucleotide metabolism. The selected LC approach, hydrophilic interaction chromatography with an amino column, effectively separates highly water soluble metabolites that fail to retain using standard reversed-phase chromatography. MS/MS detection is achieved by scanning through numerous selected reaction monitoring events on a triple quadrupole instrument. When applied to extracts of Escherichia coli grown in [12C]- versus [13C]glucose, the method reveals appropriate 12C- and 13C-peaks for 79 different metabolites. © 2006 Elsevier B.V. All rights reserved. Keywords: LC–ESI-MS/MS; Escherichia coli; Metabolomics; Metabonomics; Hydrophilic interaction chromatography; Bacteria; Metabolite; metabolism; Triple quadrupole; Mass spectrometry 1. Introduction pounds, likely capture most key metabolic end products and intermediates [5–8]. Thus, a key current need is an assay that The ability to quantify numerous mRNA in parallel using can reliably and efficiently measure these known compounds DNA microarrays has revolutionized biological science, with [9].