Khristianthesisresubmit.Pdf (7.238Mb)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

This Article Appeared in a Journal Published by Elsevier. the Attached
This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues. Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited. In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier’s archiving and manuscript policies are encouraged to visit: http://www.elsevier.com/copyright Author's personal copy ARTICLE IN PRESS European Journal of PROTISTOLOGY European Journal of Protistology 44 (2008) 299–307 www.elsevier.de/ejop Morphology and molecular phylogeny of Haplozoon praxillellae n. sp. (Dinoflagellata): A novel intestinal parasite of the maldanid polychaete Praxillella pacifica Berkeley Sonja RueckertÃ, Brian S. Leander Canadian Institute for Advanced Research, Program in Integrated Microbial Biodiversity, Departments of Botany and Zoology, University of British Columbia, Vancouver, BC, Canada V6T 1Z4 Received 11 December 2007; received in revised form 3 April 2008; accepted 5 April 2008 Abstract The genus Haplozoon comprises a group of endoparasites infecting the intestines of polychaete worms. Comparative studies using light microscopy, scanning and transmission electron microscopy, and small subunit rDNA have shown that these organisms are very unusual dinoflagellates. To date, there is only one species known from the Pacific Ocean, namely Haplozoon axiothellae Siebert. In this study, we describe Haplozoon praxillellae n. sp. from the intestine of the Pacific maldanid polychaete Praxillella pacifica Berkeley. -
Molecular Data and the Evolutionary History of Dinoflagellates by Juan Fernando Saldarriaga Echavarria Diplom, Ruprecht-Karls-Un
Molecular data and the evolutionary history of dinoflagellates by Juan Fernando Saldarriaga Echavarria Diplom, Ruprecht-Karls-Universitat Heidelberg, 1993 A THESIS SUBMITTED IN PARTIAL FULFILMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY in THE FACULTY OF GRADUATE STUDIES Department of Botany We accept this thesis as conforming to the required standard THE UNIVERSITY OF BRITISH COLUMBIA November 2003 © Juan Fernando Saldarriaga Echavarria, 2003 ABSTRACT New sequences of ribosomal and protein genes were combined with available morphological and paleontological data to produce a phylogenetic framework for dinoflagellates. The evolutionary history of some of the major morphological features of the group was then investigated in the light of that framework. Phylogenetic trees of dinoflagellates based on the small subunit ribosomal RNA gene (SSU) are generally poorly resolved but include many well- supported clades, and while combined analyses of SSU and LSU (large subunit ribosomal RNA) improve the support for several nodes, they are still generally unsatisfactory. Protein-gene based trees lack the degree of species representation necessary for meaningful in-group phylogenetic analyses, but do provide important insights to the phylogenetic position of dinoflagellates as a whole and on the identity of their close relatives. Molecular data agree with paleontology in suggesting an early evolutionary radiation of the group, but whereas paleontological data include only taxa with fossilizable cysts, the new data examined here establish that this radiation event included all dinokaryotic lineages, including athecate forms. Plastids were lost and replaced many times in dinoflagellates, a situation entirely unique for this group. Histones could well have been lost earlier in the lineage than previously assumed. -

Patrons De Biodiversité À L'échelle Globale Chez Les Dinoflagellés
! ! ! ! ! !"#$%&'%&'()!(*+!&'%&,-./01%*$0!2&30%**%&%!&4+*0%&).*0%& ! 0$'1&2(&3'!4!5&6(67&)!#2%&8)!9!:16()!;6136%2()!;&<)%=&3'!>?!@&<283! ! A%'=)83')!$2%! 45&/678&,9&:9;<6=! ! A6?% 6B3)8&% ()!7%2>) >) '()!%.*&>9&?-./01%*$0!2&30%**%&%!&4+*0%&).*0%! ! ! 0?C)3!>)!(2!3DE=)!4! ! @!!"#$%&'()*(+,%),-*$',#.(/(01.23*00*(40%+"0*(23*5(0*'( >A86B?7C9??D;&E?78<=68AFG9;&H7IA8;! ! ! ! 06?3)8?)!()!4!.+!FGH0!*+./! ! ;)<283!?8!C?%I!16#$6='!>)!4! ! 'I5&*6J987&$=9I8J!0&%!G(&=3)%!K2%>I!L6?8>23&68!M6%!N1)28!01&)81)!O0GKLN0PJ!A(I#6?3D!Q!H6I2?#)RS8&!! !!H2$$6%3)?%! 3I6B5&K78&37J?6J;LAJ!S8&<)%=&3'!>)!T)8E<)!Q!0?&==)! !!H2$$6%3)?%! 'I5&47IA87&468=I9;6IJ!032U&68)!V66(67&12!G8368!;6D%8!6M!W2$()=!Q!"32(&)! XY2#&823)?%! 3I6B5&,7I;&$=9HH788J!SAFZ,ZWH0!0323&68!V66(67&[?)!>)!@&(()M%281D)R=?%RF)%!Q!L%281)! XY2#&823)?%! 'I5&*7BB79?9&$A786J!;\WXZN,A)(276=J!"LHXFXH!!"#$%"&'"&(%")$*&+,-./0#1&Q!L%281)!!! !!!Z6R>&%)13)?%!>)!3DE=)! 'I5&)6?6HM78&>9&17IC7;J&SAFZ,ZWH0!0323&68!5&6(67&[?)!>)!H6=16MM!Q!L%281)! ! !!!!!!!!!;&%)13)?%!>)!3DE=)! ! ! ! "#$%&#'!()!*+,+-,*+./! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! Remerciements* ! Remerciements* A!l'issue!de!ce!travail!de!recherche!et!de!sa!rédaction,!j’ai!la!preuve!que!la!thèse!est!loin!d'être!un!travail! solitaire.! En! effet,! je! n'aurais! jamais! pu! réaliser! ce! travail! doctoral! sans! le! soutien! d'un! grand! nombre! de! personnes!dont!l’amitié,!la!générosité,!la!bonne!humeur%et%l'intérêt%manifestés%à%l'égard%de%ma%recherche%m'ont% permis!de!progresser!dans!cette!phase!délicate!de!«!l'apprentiGchercheur!».! -

A Parasite of Marine Rotifers: a New Lineage of Dinokaryotic Dinoflagellates (Dinophyceae)
Hindawi Publishing Corporation Journal of Marine Biology Volume 2015, Article ID 614609, 5 pages http://dx.doi.org/10.1155/2015/614609 Research Article A Parasite of Marine Rotifers: A New Lineage of Dinokaryotic Dinoflagellates (Dinophyceae) Fernando Gómez1 and Alf Skovgaard2 1 Laboratory of Plankton Systems, Oceanographic Institute, University of Sao˜ Paulo, Prac¸a do Oceanografico´ 191, Cidade Universitaria,´ 05508-900 Butanta,˜ SP, Brazil 2Department of Veterinary Disease Biology, University of Copenhagen, Stigbøjlen 7, 1870 Frederiksberg C, Denmark Correspondence should be addressed to Fernando Gomez;´ [email protected] Received 11 July 2015; Accepted 27 August 2015 Academic Editor: Gerardo R. Vasta Copyright © 2015 F. Gomez´ and A. Skovgaard. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Dinoflagellate infections have been reported for different protistan and animal hosts. We report, for the first time, the association between a dinoflagellate parasite and a rotifer host, tentatively Synchaeta sp. (Rotifera), collected from the port of Valencia, NW Mediterranean Sea. The rotifer contained a sporangium with 100–200 thecate dinospores that develop synchronically through palintomic sporogenesis. This undescribed dinoflagellate forms a new and divergent fast-evolved lineage that branches amongthe dinokaryotic dinoflagellates. 1. Introduction form independent lineages with no evident relation to other dinoflagellates [12]. In this study, we describe a new lineage of The alveolates (or Alveolata) are a major lineage of protists an undescribed parasitic dinoflagellate that largely diverged divided into three main phyla: ciliates, apicomplexans, and from other known dinoflagellates. -
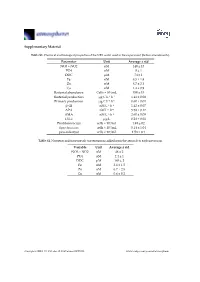
Supplementary Material Parameter Unit Average ± Std NO3 + NO2 Nm
Supplementary Material Table S1. Chemical and biological properties of the NRS water used in the experiment (before amendments). Parameter Unit Average ± std NO3 + NO2 nM 140 ± 13 PO4 nM 8 ± 1 DOC μM 74 ± 1 Fe nM 8.5 ± 1.8 Zn nM 8.7 ± 2.1 Cu nM 1.4 ± 0.9 Bacterial abundance Cells × 104/mL 350 ± 15 Bacterial production μg C L−1 h−1 1.41 ± 0.08 Primary production μg C L−1 h−1 0.60 ± 0.01 β-Gl nM L−1 h−1 1.42 ± 0.07 APA nM L−1 h−1 5.58 ± 0.17 AMA nM L−1·h−1 2.60 ± 0.09 Chl-a μg/L 0.28 ± 0.01 Prochlorococcus cells × 104/mL 1.49 ± 02 Synechococcus cells × 104/mL 5.14 ± 1.04 pico-eukaryot cells × 103/mL 1.58 × 0.1 Table S2. Nutrients and trace metals concentrations added from the aerosols to each mesocosm. Variable Unit Average ± std NO3 + NO2 nM 48 ± 2 PO4 nM 2.4 ± 1 DOC μM 165 ± 2 Fe nM 2.6 ± 1.5 Zn nM 6.7 ± 2.5 Cu nM 0.6 ± 0.2 Atmosphere 2019, 10, 358; doi:10.3390/atmos10070358 www.mdpi.com/journal/atmosphere Atmosphere 2019, 10, 358 2 of 6 Table S3. ANOVA test results between control, ‘UV-treated’ and ‘live-dust’ treatments at 20 h or 44 h, with significantly different values shown in bold. ANOVA df Sum Sq Mean Sq F Value p-value Chl-a 20 H 2, 6 0.03, 0.02 0.02, 0 4.52 0.0634 44 H 2, 6 0.02, 0 0.01, 0 23.13 0.002 Synechococcus Abundance 20 H 2, 7 8.23 × 107, 4.11 × 107 4.11 × 107, 4.51 × 107 0.91 0.4509 44 H 2, 7 5.31 × 108, 6.97 × 107 2.65 × 108, 1.16 × 107 22.84 0.0016 Prochlorococcus Abundance 20 H 2, 8 4.22 × 107, 2.11 × 107 2.11 × 107, 2.71 × 106 7.77 0.0216 44 H 2, 8 9.02 × 107, 1.47 × 107 4.51 × 107, 2.45 × 106 18.38 0.0028 Pico-eukaryote -

Protist Phylogeny and the High-Level Classification of Protozoa
Europ. J. Protistol. 39, 338–348 (2003) © Urban & Fischer Verlag http://www.urbanfischer.de/journals/ejp Protist phylogeny and the high-level classification of Protozoa Thomas Cavalier-Smith Department of Zoology, University of Oxford, South Parks Road, Oxford, OX1 3PS, UK; E-mail: [email protected] Received 1 September 2003; 29 September 2003. Accepted: 29 September 2003 Protist large-scale phylogeny is briefly reviewed and a revised higher classification of the kingdom Pro- tozoa into 11 phyla presented. Complementary gene fusions reveal a fundamental bifurcation among eu- karyotes between two major clades: the ancestrally uniciliate (often unicentriolar) unikonts and the an- cestrally biciliate bikonts, which undergo ciliary transformation by converting a younger anterior cilium into a dissimilar older posterior cilium. Unikonts comprise the ancestrally unikont protozoan phylum Amoebozoa and the opisthokonts (kingdom Animalia, phylum Choanozoa, their sisters or ancestors; and kingdom Fungi). They share a derived triple-gene fusion, absent from bikonts. Bikonts contrastingly share a derived gene fusion between dihydrofolate reductase and thymidylate synthase and include plants and all other protists, comprising the protozoan infrakingdoms Rhizaria [phyla Cercozoa and Re- taria (Radiozoa, Foraminifera)] and Excavata (phyla Loukozoa, Metamonada, Euglenozoa, Percolozoa), plus the kingdom Plantae [Viridaeplantae, Rhodophyta (sisters); Glaucophyta], the chromalveolate clade, and the protozoan phylum Apusozoa (Thecomonadea, Diphylleida). Chromalveolates comprise kingdom Chromista (Cryptista, Heterokonta, Haptophyta) and the protozoan infrakingdom Alveolata [phyla Cilio- phora and Miozoa (= Protalveolata, Dinozoa, Apicomplexa)], which diverged from a common ancestor that enslaved a red alga and evolved novel plastid protein-targeting machinery via the host rough ER and the enslaved algal plasma membrane (periplastid membrane). -

Ellobiopsids of the Genus Thalassomyces Are Alveolates
J. Eukaryot. Microbiol., 51(2), 2004 pp. 246±252 q 2004 by the Society of Protozoologists Ellobiopsids of the Genus Thalassomyces are Alveolates JEFFREY D. SILBERMAN,a,b1 ALLEN G. COLLINS,c,2 LISA-ANN GERSHWIN,d,3 PATRICIA J. JOHNSONa and ANDREW J. ROGERe aDepartment of Microbiology, Immunology, and Molecular Genetics, University of California at Los Angeles, California, USA, and bInstitute of Geophysics and Planetary Physics, University of California at Los Angeles, California, USA, and cEcology, Behavior and Evolution Section, Division of Biology, University of California, La Jolla, California, USA, and dDepartment of Integrative Biology and Museum of Paleontology, University of California, Berkeley, California, USA, and eCanadian Institute for Advanced Research, Program in Evolutionary Biology, Genome Atlantic, Department of Biochemistry and Molecular Biology, Dalhousie University, Halifax, Nova Scotia, Canada ABSTRACT. Ellobiopsids are multinucleate protist parasites of aquatic crustaceans that possess a nutrient absorbing `root' inside the host and reproductive structures that protrude through the carapace. Ellobiopsids have variously been af®liated with fungi, `colorless algae', and dino¯agellates, although no morphological character has been identi®ed that de®nitively allies them with any particular eukaryotic lineage. The arrangement of the trailing and circumferential ¯agella of the rarely observed bi-¯agellated `zoospore' is reminiscent of dino¯agellate ¯agellation, but a well-organized `dinokaryotic nucleus' has never been observed. Using small subunit ribosomal RNA gene sequences from two species of Thalassomyces, phylogenetic analyses robustly place these ellobiopsid species among the alveolates (ciliates, apicomplexans, dino¯agellates and relatives) though without a clear af®liation to any established alveolate lineage. Our trees demonstrate that Thalassomyces fall within a dino¯agellate 1 apicomplexa 1 Perkinsidae 1 ``marine alveolate group 1'' clade, clustering most closely with dino¯agellates. -

Dinoflagellate Nuclear SSU Rrna Phylogeny Suggests Multiple Plastid Losses and Replacements
J Mol Evol (2001) 53:204–213 DOI: 10.1007/s002390010210 © Springer-Verlag New York Inc. 2001 Dinoflagellate Nuclear SSU rRNA Phylogeny Suggests Multiple Plastid Losses and Replacements Juan F. Saldarriaga,1 F.J.R. Taylor,1,2 Patrick J. Keeling,1 Thomas Cavalier-Smith3 1 Department of Botany, University of British Columbia, 6270 University Boulevard, Vancouver, British Columbia, V6T 1Z4, Canada 2 Department of Earth and Ocean Sciences, University of British Columbia, 6270 University Boulevard, Vancouver, British Columbia, V6T 1Z4, Canada 3 Department of Zoology, Oxford University, South Parks Road, Oxford, OX1 3PS, UK Received: 25 September 2000 / Accepted: 24 April 2001 Abstract. Dinoflagellates are a trophically diverse Introduction group of protists with photosynthetic and non- photosynthetic members that appears to incorporate and There is now no serious doubt that mitochondria and lose endosymbionts relatively easily. To trace the gain plastids are descendants of free-living prokaryotic cells and loss of plastids in dinoflagellates, we have sequenced (Gray and Spencer 1996). The primary endosymbioses the nuclear small subunit rRNA gene of 28 photosyn- that incorporated these cells into eukaryotic organisms thetic and four non-photosynthetic species, and produced are, however, exceedingly rare events: mitochondria phylogenetic trees with a total of 81 dinoflagellate se- were probably incorporated only once in the history of quences. Patterns of plastid gain, loss, and replacement life (Roger 1999), and the same is probably true for were plotted onto this phylogeny. With the exception of plastids (Delwiche 1999; Cavalier-Smith 2000). Vertical the apparently early-diverging Syndiniales and Noctilu- descendants of plastids obtained through primary endo- cales, all non-photosynthetic dinoflagellates are very symbiosis are now found in many photosynthetic organ- likely to have had photosynthetic ancestors with peridi- isms (glaucophytes, red and green algae, and land nin-containing plastids. -

Redalyc.Paratrichodina Africana (Ciliophora: Trichodinidae) of Wild and Cultured Nile Tilapia in the Northern Brazil
Revista Brasileira de Parasitologia Veterinária ISSN: 0103-846X [email protected] Colégio Brasileiro de Parasitologia Veterinária Brasil Tavares-Dias, Marcos; da Costa Marchiori, Natália; Laterça Martins, Maurício Paratrichodina africana (Ciliophora: Trichodinidae) of wild and cultured Nile tilapia in the Northern Brazil Revista Brasileira de Parasitologia Veterinária, vol. 22, núm. 2, abril-junio, 2013, pp. 248- 252 Colégio Brasileiro de Parasitologia Veterinária Jaboticabal, Brasil Available in: http://www.redalyc.org/articulo.oa?id=397841488011 How to cite Complete issue Scientific Information System More information about this article Network of Scientific Journals from Latin America, the Caribbean, Spain and Portugal Journal's homepage in redalyc.org Non-profit academic project, developed under the open access initiative Full Article Rev. Bras. Parasitol. Vet., Jaboticabal, v. 22, n. 2, p. 248-252, abr.-jun. 2013 ISSN 0103-846X (impresso) / ISSN 1984-2961 (eletrônico) Paratrichodina africana (Ciliophora: Trichodinidae) of wild and cultured Nile tilapia in the Northern Brazil Paratrichodina africana (Ciliophora: Trichodinidae) de tilápia do Nilo selvagem e cultivada no Norte do Brasil Marcos Tavares-Dias1; Natália da Costa Marchiori2; Maurício Laterça Martins2* 1Laboratório de Aquicultura e Pesca, Embrapa Amapá, Macapá, AP, Brasil 2Laboratório de Sanidade em Organismos Aquáticos – AQUOS, Departamento de Aquicultura, Universidade Federal de Santa Catarina – UFSC, Florianópolis, SC, Brasil Received September 8, 2012 Accepted February 1, 2013 Abstract The present work morphologically characterizes Paratrichodina africana from the gills of wild and farmed Nile tilapia from Northern Brazil (eastern Amazonia). Ninety fish were captured for parasitological analysis in Macapá, State of Amapá, from a wetland area bathed by the Amazon River commonly called ‘Ressaca do Zerão’ (n = 52), as well as from a local fish farm (n = 38). -

D070p001.Pdf
DISEASES OF AQUATIC ORGANISMS Vol. 70: 1–36, 2006 Published June 12 Dis Aquat Org OPENPEN ACCESSCCESS FEATURE ARTICLE: REVIEW Guide to the identification of fish protozoan and metazoan parasites in stained tissue sections D. W. Bruno1,*, B. Nowak2, D. G. Elliott3 1FRS Marine Laboratory, PO Box 101, 375 Victoria Road, Aberdeen AB11 9DB, UK 2School of Aquaculture, Tasmanian Aquaculture and Fisheries Institute, CRC Aquafin, University of Tasmania, Locked Bag 1370, Launceston, Tasmania 7250, Australia 3Western Fisheries Research Center, US Geological Survey/Biological Resources Discipline, 6505 N.E. 65th Street, Seattle, Washington 98115, USA ABSTRACT: The identification of protozoan and metazoan parasites is traditionally carried out using a series of classical keys based upon the morphology of the whole organism. However, in stained tis- sue sections prepared for light microscopy, taxonomic features will be missing, thus making parasite identification difficult. This work highlights the characteristic features of representative parasites in tissue sections to aid identification. The parasite examples discussed are derived from species af- fecting finfish, and predominantly include parasites associated with disease or those commonly observed as incidental findings in disease diagnostic cases. Emphasis is on protozoan and small metazoan parasites (such as Myxosporidia) because these are the organisms most likely to be missed or mis-diagnosed during gross examination. Figures are presented in colour to assist biologists and veterinarians who are required to assess host/parasite interactions by light microscopy. KEY WORDS: Identification · Light microscopy · Metazoa · Protozoa · Staining · Tissue sections Resale or republication not permitted without written consent of the publisher INTRODUCTION identifying the type of epithelial cells that compose the intestine. -

Marinha Do Brasil Instituto De Estudos Do Mar Almirante Paulo Moreira Universidade Federal Fluminense Programa Associado De Pós-Graduação Em Biotecnologia Marinha
MARINHA DO BRASIL INSTITUTO DE ESTUDOS DO MAR ALMIRANTE PAULO MOREIRA UNIVERSIDADE FEDERAL FLUMINENSE PROGRAMA ASSOCIADO DE PÓS-GRADUAÇÃO EM BIOTECNOLOGIA MARINHA LAIS PEREIRA D' OLIVEIRA NAVAL XAVIER ECO-ENGENHARIA EM ÁREA PORTUÁRIA E SUAS IMPLICAÇÕES NA BIOINCRUSTAÇÃO E NA PREVENÇÃO DOS PROCESSOS DE BIOINVASÃO. ARRAIAL DO CABO 2018 MARINHA DO BRASIL INSTITUTO DE ESTUDOS DO MAR ALMIRANTE PAULO MOREIRA UNIVERSIDADE FEDERAL FLUMINENSE PROGRAMA ASSOCIADO DE PÓS-GRADUAÇÃO EM BIOTECNOLOGIA MARINHA LAIS PEREIRA D' OLIVEIRA NAVAL XAVIER ECO-ENGENHARIA EM ÁREA PORTUÁRIA E SUAS IMPLICAÇÕES NA BIOINCRUSTAÇÃO E NA PREVENÇÃO DOS PROCESSOS DE BIOINVASÃO. Dissertação de Mestrado apresentada ao Instituto de Estudos do Mar Almirante Paulo Moreira e à Universidade Federal Fluminense, como requisito parcial para a obtenção do grau de Mestre em Biotecnologia Marinha. Orientador: Dr. Ricardo Coutinho Coorientadora: Dra. Luciana V.R. de Messano ARRAIAL DO CABO 2018 LAIS PEREIRA D' OLIVEIRA NAVAL XAVIER ECO-ENGENHARIA EM ÁREA PORTUÁRIA E SUAS IMPLICAÇÕES NA BIOINCRUSTAÇÃO E NA PREVENÇÃO DOS PROCESSOS DE BIOINVASÃO. Dissertação apresentada ao Instituto de Estudos do Mar Almirante Paulo Moreira e à Universidade Federal Fluminense, como requisito parcial para a obtenção do título de Mestre em Biotecnologia Marinha. COMISSÃO JULGADORA: ___________________________________________________________________ Dr. Ricardo Coutinho Instituto de Estudos do Mar Almirante Paulo Moreira Professor orientador - Presidente da Banca Examinadora ___________________________________________________________________ -

Spatio-Temporal Distribution, Physiological Characterization and Toxicity of the Marine Dinoflagellate Ostreopsis (Schmidt) from a Temperate Area, the Ebre Delta
Spatio-temporal distribution, physiological characterization and toxicity of the marine dinoflagellate Ostreopsis (Schmidt) from a temperate area, the Ebre Delta. Phylogenetic variability in comparison with a tropical area, Reunion Island Olga Carnicer Castaño ADVERTIMENT. La consulta d’aquesta tesi queda condicionada a l’acceptació de les següents condicions d'ús: La difusió d’aquesta tesi per mitjà del servei TDX (www.tdx.cat) i a través del Dipòsit Digital de la UB (diposit.ub.edu) ha estat autoritzada pels titulars dels drets de propietat intel·lectual únicament per a usos privats emmarcats en activitats d’investigació i docència. No s’autoritza la seva reproducció amb finalitats de lucre ni la seva difusió i posada a disposició des d’un lloc aliè al servei TDX ni al Dipòsit Digital de la UB. No s’autoritza la presentació del seu contingut en una finestra o marc aliè a TDX o al Dipòsit Digital de la UB (framing). Aquesta reserva de drets afecta tant al resum de presentació de la tesi com als seus continguts. En la utilització o cita de parts de la tesi és obligat indicar el nom de la persona autora. ADVERTENCIA. La consulta de esta tesis queda condicionada a la aceptación de las siguientes condiciones de uso: La difusión de esta tesis por medio del servicio TDR (www.tdx.cat) y a través del Repositorio Digital de la UB (diposit.ub.edu) ha sido autorizada por los titulares de los derechos de propiedad intelectual únicamente para usos privados enmarcados en actividades de investigación y docencia. No se autoriza su reproducción con finalidades de lucro ni su difusión y puesta a disposición desde un sitio ajeno al servicio TDR o al Repositorio Digital de la UB.