Genetic Evidence for Shared Mechanisms of Epimorphic Regeneration in Zebrafish
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Notch-Signaling in Retinal Regeneration and Müller Glial Plasticity
Notch-Signaling in Retinal Regeneration and Müller glial Plasticity DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Kanika Ghai, MS Neuroscience Graduate Studies Program The Ohio State University 2009 Dissertation Committee: Dr. Andy J Fischer, Advisor Dr. Heithem El-Hodiri Dr. Susan Cole Dr. Paul Henion Copyright by Kanika Ghai 2009 ABSTRACT Eye diseases such as blindness, age-related macular degeneration (AMD), diabetic retinopathy and glaucoma are highly prevalent in the developed world, especially in a rapidly aging population. These sight-threatening diseases all involve the progressive loss of cells from the retina, the light-sensing neural tissue that lines the back of the eye. Thus, developing strategies to replace dying retinal cells or prolonging neuronal survival is essential to preserving sight. In this regard, cell-based therapies hold great potential as a treatment for retinal diseases. One strategy is to stimulate cells within the retina to produce new neurons. This dissertation elucidates the properties of the primary support cell in the chicken retina, known as the Müller glia, which have recently been shown to possess stem-cell like properties, with the potential to form new neurons in damaged retinas. However, the mechanisms that govern this stem-cell like ability are less well understood. In order to better understand these properties, we analyze the role of one of the key developmental processes, i.e., the Notch-Signaling Pathway in regulating proliferative, neuroprotective and regenerative properties of Müller glia and bestow them with this plasticity. -

Müller Glia Regenerative Potential Is Maintained Throughout Life Despite 2 Neurodegeneration and Gliosis in the Ageing Zebrafish Retina 3 4 AUTHORS 5 Raquel R
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.28.174821; this version posted October 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 Müller Glia regenerative potential is maintained throughout life despite 2 neurodegeneration and gliosis in the ageing zebrafish retina 3 4 AUTHORS 5 Raquel R. Martins1,2, Mazen Zamzam3, Mariya Moosajee4,5,6,7, Ryan Thummel3, Catarina M. 6 Henriques1,2§, Ryan B. MaCDonald4§ 7 8 Affiliations: 9 1. Bateson Centre, University of Sheffield, Sheffield, S10 2TN, UK. 10 2. Department of OnCology and Metabolism, University of Sheffield, Sheffield, S10 2TN 11 UK. 12 3. Department of Ophthalmology, Visual and AnatomiCal SCienCes, Wayne State 13 University School of MediCine, Detroit, MI 48201, USA. 14 4. Institute of Ophthalmology, University College London, London, EC1V 9EL, UK. 15 5. Moorfields Eye Hospital NHS Foundation Trust, London, EC1V 2PD, UK 16 6. Great Ormond Street Hospital for Children NHS Foundation Trust, London, WC1N 17 3JH, UK 18 7. The FranCis CriCk Institute, London, NW1 1AT, London 19 20 21 22 §Co-corresponding authors: [email protected] and [email protected] 23 24 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.06.28.174821; this version posted October 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 25 ABSTRACT 26 Ageing is a signifiCant risk faCtor for degeneration of the retina. -

Mesenchymal Stem Cell-Based Therapy for Retinal Degenerative Diseases: Experimental Models and Clinical Trials
cells Review Mesenchymal Stem Cell-Based Therapy for Retinal Degenerative Diseases: Experimental Models and Clinical Trials Vladimir Holan 1,2,*, Katerina Palacka 1,2 and Barbora Hermankova 1 1 Department of Nanotoxicology and Molecular Epidemiology, Institute of Experimental Medicine of the Czech Academy of Sciences, 14220 Prague, Czech Republic; [email protected] (K.P.); [email protected] (B.H.) 2 Department of Cell Biology, Faculty of Science, Charles University, 12843 Prague, Czech Republic * Correspondence: [email protected] Abstract: Retinal degenerative diseases, such as age-related macular degeneration, retinitis pigmen- tosa, diabetic retinopathy or glaucoma, represent the main causes of a decreased quality of vision or even blindness worldwide. However, despite considerable efforts, the treatment possibilities for these disorders remain very limited. A perspective is offered by cell therapy using mesenchymal stem cells (MSCs). These cells can be obtained from the bone marrow or adipose tissue of a particular patient, expanded in vitro and used as the autologous cells. MSCs possess potent immunoregulatory properties and can inhibit a harmful inflammatory reaction in the diseased retina. By the production of numerous growth and neurotrophic factors, they support the survival and growth of retinal cells. In addition, MSCs can protect retinal cells by antiapoptotic properties and could contribute to the regeneration of the diseased retina by their ability to differentiate into various cell types, including the cells of the retina. All of these properties indicate the potential of MSCs for the therapy of diseased Citation: Holan, V.; Palacka, K.; retinas. This view is supported by the recent results of numerous experimental studies in different Hermankova, B. -

Forging Iron Man
OCTOBER 2016 # 34 Upfront In Practice Profession Sitting Down With Keeping an eye Keratoplasty: Applying clinical trial data to The master of macular on Zika challenging convention your practice degeneration, Philip J. Rosenfeld 14 32 – 38 44 – 46 50 – 51 Forging Iron Man History is made: the story of the first clinical use of a robot in eye surgery 18 – 29 www.theophthalmologist.com ADVANCING TOGETHER 11707-Alcon-Surg-OPHTHALMOLOGIST-OCTOBER-ISSUE-OUTLINED.indd 1 05/09/2016 13:49 Image of the ADVANCING Month TOGETHER Confetti Cornea A cornea from a K14CreER-Confetti mouse containing the four color Brainbow reporter cassette. The multicolored radial streaks develop after induction of the transgene with tamoxifen, and arise from Keratin 14-expressing progenitor cells, positioned in the limbal annulus. Nick Di Girolamo and his colleagues developed the model to better understand basic corneal biology and how stem cells function to replenish the cornea throughout life. Nick says “This model lends itself beautifully to studying when corneal stem cells are designated, their destiny during aging, and how they behave during corneal wounding and following transplantation. We believe this technology will be used to help address some of the controversies and limitations that have plagued our field for decades. Our ultimate goal is to translate our findings to the clinic.” Image courtesy of Associate Professor Nick Di Girolamo from the School of Medical Science, University of New South Wales, Sydney, Australia. Do you have an image you’d like to see featured in The Ophthalmologist? Contact [email protected]. www.theophthalmologist.com 11707-Alcon-Surg-OPHTHALMOLOGIST-OCTOBER-ISSUE-OUTLINED.indd 1 05/09/2016 13:49 Contents 14 In My View 16 Robert Ritch puts forward the case for iridoplasty as an effective means of opening an appositionally closed angle, and 18 shares his experiences with the technique. -

Regeneration of Inner Retinal Neurons After Intravitreal Injection of Ouabain in Zebrafish
1712 • The Journal of Neuroscience, February 14, 2007 • 27(7):1712–1724 Development/Plasticity/Repair Regeneration of Inner Retinal Neurons after Intravitreal Injection of Ouabain in Zebrafish Shane M. Fimbel, Jacob E. Montgomery, Christopher T. Burket, and David R. Hyde Department of Biological Sciences and Center for Zebrafish Research, University of Notre Dame, Notre Dame, Indiana 46556 We examined the regenerative capacity of the adult zebrafish retina by intravitreal injection of a low ouabain concentration to rapidly damage the ganglion cell layer (GCL) and inner nuclear layer (INL) with minimal photoreceptor cell damage. By 24 h after ouabain injection, maximal numbers of terminal deoxynucleotidyl transferase-mediated biotinylated UTP nick end labeling (TUNEL)-positive cells were detected in the INL and GCL, with low numbers of TUNEL-positive cells in the outer nuclear layer. Immunolabeling revealed that ϳ85% of the HuC/D-positive amacrine and ganglion cells were lost by 7 d post-ouabain injection (dpi). This ganglion cell loss was consistent with the small, but statistically significant, decrease in the optic nerve diameter. The regeneration response began within 1 dpi with increased proliferating cell nuclear antigen (PCNA) expression in both the INL and GCL. By 3 dpi, PCNA expression is primarily restricted to the Mu¨ller glia. By 5 dpi, most of the PCNA expression was localized to neuronal progenitors expressing the olig2:egfp transgene rather than the Mu¨ller glia. By 7 dpi, the neuronal progenitors began committing to the ganglion cell fate based on the coexpression of the atoh7:EGFP transgene and the zn5 antigen. The regeneration of ganglion and amacrine cells continued until 60 dpi, when they reached 75% of their uninjected control number. -

Wnt Signaling Promotes Regeneration in the Retina of Adult Mammals
4210 • The Journal of Neuroscience, April 11, 2007 • 27(15):4210–4219 Development/Plasticity/Repair Wnt Signaling Promotes Regeneration in the Retina of Adult Mammals Fumitaka Osakada,1,2,3* Sotaro Ooto,4* Tadamichi Akagi,4 Michiko Mandai,1 Akinori Akaike,3 and Masayo Takahashi1,2 1Laboratory for Retinal Regeneration, Center for Developmental Biology, RIKEN, Kobe 650-0047, Japan, 2Department of Experimental Therapeutics, Translational Research Center, Kyoto University Hospital, Kyoto 606-8507, Japan, 3Department of Pharmacology, Graduate School of Pharmaceutical Sciences, Kyoto University, Kyoto 606-8501, Japan, and 4Department of Ophthalmology and Visual Sciences, Graduate School of Medicine, Kyoto University, Kyoto 606-8507, Japan Regeneration in the mammalian CNS is severely limited. Unlike in the chick, current models hold that retinal neurons are never regen- erated. Previously we demonstrated that, in the adult mammalian retina, Mu¨ller glia dedifferentiate and produce retinal cells, including photoreceptors, after acute neurotoxic injury in vivo. However, the number of newly generated retinal neurons is very limited. Here we demonstrate that Wnt (wingless-type MMTV integration site family)/-catenin signaling promotes proliferation of Mu¨ller glia-derived retinal progenitors and neural regeneration after damage or during degeneration. Wnt3a treatment increases proliferation of dediffer- entiated Mu¨ller glia Ͼ20-fold in the photoreceptor-damaged retina. Supplementation with retinoic acid or valproic acid induces differ- entiation of these cells primarily into Crx (cone rod homeobox)-positive and rhodopsin-positive photoreceptors. Notably, injury induces nuclear accumulation of -catenin, cyclin D1 upregulation, and Wnt/-catenin reporter activity. Activation of Wnt signaling by glycogen synthase kinase-3 inhibitors promotes retinal regeneration, and, conversely, inhibition of the signaling attenuates regeneration. -
Restoring Vision to the Blind the Lasker/IRRF Initiative for Innovation in Vision Science
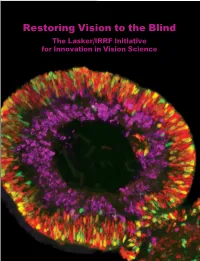
Restoring Vision to the Blind The Lasker/IRRF Initiative for Innovation in Vision Science About the Albert and Mary Lasker Foundation: Founded in 1942, the Albert and Mary Lasker Foundation envisions a healthier world through sustained support for basic and clinical medical research. The Foundation works to accomplish its mission through education and advocacy and, most notably, through a prestigious annual awards program, now in its 70th year. Lasker Award winners are selected by their peers, who, like themselves, include the world’s most accomplished and well-respected Cover: Optic vesicle-like structures derived from human induced pluripotent stem cells can self medical research scientists, and thus the award represents a special honor. The Foundation’s education assemble into rudimentary retinal laminae, and following 50 days of differentiation, form an outer and advocacy missions focus on engaging the public and policymakers on the importance of robust neuroblastic layer of proliferating progenitor cells (identified by immunolabeling with the retinal medical research programs and the funding to make them possible. The Lasker Foundation is also progenitor marker VSX2, in red, and the mitotic cell marker Ki67, in green) and an inner layer dedicated to supporting and inspiring the next generation of research scientists. For more information of putative retinal ganglion cells (immunolabeled with HuCD, a marker for post-mitotic neurons, about the Lasker Foundation and its programs, visit http://www.laskerfoundation.org. in purple). Image produced by the Gamm Laboratory, University of Wisconsin School of Medicine and Public Health. Reprinted from: Wright LS, Phillips MJ, Pinilla I, Hei D, Gamm D. Induced pluripotent stem cells as custom therapeutics for retinal repair: Progress and rationale. -

Retinal Stem Cell ‘Retirement Plans’: Growth, Regulation and Species Adaptations in the Retinal Ciliary Marginal Zone
International Journal of Molecular Sciences Review Retinal Stem Cell ‘Retirement Plans’: Growth, Regulation and Species Adaptations in the Retinal Ciliary Marginal Zone Amanda Miles and Vincent Tropepe * Department of Cell & Systems Biology, University of Toronto, 25 Harbord Street, Toronto, ON M5S 3G5, Canada; [email protected] * Correspondence: [email protected]; Tel.: +416-9460338 Abstract: The vertebrate retina develops from a specified group of precursor cells that adopt distinct identities and generate lineages of either the neural retina, retinal pigmented epithelium, or ciliary body. In some species, including teleost fish and amphibians, proliferative cells with stem-cell- like properties capable of continuously supplying new retinal cells post-embryonically have been characterized and extensively studied. This region, termed the ciliary or circumferential marginal zone (CMZ), possibly represents a conserved retinal stem cell niche. In this review, we highlight the research characterizing similar CMZ-like regions, or stem-like cells located at the peripheral margin, across multiple different species. We discuss the proliferative parameters, multipotency and growth mechanisms of these cells to understand how they behave in vivo and how different molecular factors and signalling networks converge at the CMZ niche to regulate their activity. The evidence suggests that the mature retina may have a conserved propensity for homeostatic growth and plasticity and that dysfunction in the regulation of CMZ activity may partially account for dystrophic eye growth Citation: Miles, A.; Tropepe, V. diseases such as myopia and hyperopia. A better understanding of the properties of CMZ cells will Retinal Stem Cell ‘Retirement Plans’: enable important insight into how an endogenous generative tissue compartment can adapt to altered Growth, Regulation and Species retinal physiology and potentially even restore vision loss caused by retinal degenerative conditions. -

(Her) Genes During Vertebrate Retinal Development and Regeneration
University of Kentucky UKnowledge Theses and Dissertations--Biology Biology 2016 ROLE OF HAIRY-RELATED (HER) GENES DURING VERTEBRATE RETINAL DEVELOPMENT AND REGENERATION Stephen G. Wilson University of Kentucky, [email protected] Digital Object Identifier: http://dx.doi.org/10.13023/ETD.2016.249 Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation Wilson, Stephen G., "ROLE OF HAIRY-RELATED (HER) GENES DURING VERTEBRATE RETINAL DEVELOPMENT AND REGENERATION" (2016). Theses and Dissertations--Biology. 36. https://uknowledge.uky.edu/biology_etds/36 This Doctoral Dissertation is brought to you for free and open access by the Biology at UKnowledge. It has been accepted for inclusion in Theses and Dissertations--Biology by an authorized administrator of UKnowledge. For more information, please contact [email protected]. STUDENT AGREEMENT: I represent that my thesis or dissertation and abstract are my original work. Proper attribution has been given to all outside sources. I understand that I am solely responsible for obtaining any needed copyright permissions. I have obtained needed written permission statement(s) from the owner(s) of each third-party copyrighted matter to be included in my work, allowing electronic distribution (if such use is not permitted by the fair use doctrine) which will be submitted to UKnowledge as Additional File. I hereby grant to The University of Kentucky and its agents the irrevocable, non-exclusive, and royalty-free license to archive and make accessible my work in whole or in part in all forms of media, now or hereafter known. -

Insights on the Regeneration Potential of Müller Glia in the Mammalian Retina
cells Review Insights on the Regeneration Potential of Müller Glia in the Mammalian Retina Ahmed Salman 1,* , Michelle E. McClements 1 and Robert E. MacLaren 1,2 Citation: Salman, A.; McClements, M.E.; MacLaren, R.E. Insights on the Regeneration Potential of Müller Glia in the Mammalian Retina. Cells 2021, 10, 1957. https://doi.org/10.3390/ cells10081957 Academic Editor: Alexander E. Kalyuzhny Received: 19 June 2021 Accepted: 26 July 2021 Published: 31 July 2021 Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affil- iations. Copyright: © 2021 by the authors. Cells 2021, 10, 1957 2 of 12 systems for unknown reasons. Gaining more understanding of the mechanisms of the re- generation potential in fish may help unlock the reasons why mammalian cells lack this phenomenon and how we might encourage it. This review will consider the responses of Müller glia to retinal injury in mammals and the signalling mechanisms underlying their reprogramming and regeneration poten- tial. 2. Müller Glia Anatomy and Function Cells 2021, 10, 1957 The function of the retina is to convey light signals to the brain. The photoreceptor 2 of 12 cells respond to light, triggering a phototransduction cascade that passes through the bi- polar cells, then on to the retinal ganglion cells that converge into the optic nerve, which carries the signalregeneration to the brain. potential Horizontal in fish cells may enable help interconnections unlock the reasons of whymultiple mammalian photo- cells lack this receptors and bipolarphenomenon cells, whilst and how amacrine we might cells encourage are the interneurons it. -

Cellular Strategies for Retinal Repair by Photoreceptor Replacement
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by UCL Discovery Jayakody, SA; Gonzalez-Cordero, A; Ali, RR; Pearson, RA; (2015) Cellular strategies for retinal repair by photoreceptor replacement. Prog Retin Eye Res , 46 31 - 66. 10.1016/j.preteyeres.2015.01.003. ARTICLE Cellular strategies for retinal repair by photoreceptor replacement Sujatha A. Jayakody1, Anai Gonzalez-Cordero1, Robin R Ali1,2,, Rachael A. Pearson1$ 1 Gene and Cell Therapy Group, Department of Genetics, UCL Institute of Ophthalmology, 11-43 Bath St, London EC1V 9EL, UK 2 NIHR Biomedical Research Centre at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology, City Road, London EC1V 2PD, UK Corresponding author: $ Correspondence should be sent to Dr Rachael A. Pearson, Department of Genetics, University College London Institute of Ophthalmology, 11-43 Bath Street, London, EC1V 9EL, +44 (0) 2076084022, [email protected] http://www.ucl.ac.uk/ioo/research/ali.htm Abstract: Loss of photoreceptors due to retinal degeneration is a major cause of blindness in the developed world. While no effective treatment is currently available, cell replacement therapy, using pluripotent stem cell-derived photoreceptor precursor cells, may be a feasible future treatment. Recent reports have demonstrated rescue of visual function following the transplantation of immature photoreceptors and we have seen major advances in our ability to generate transplantation-competent donor cells from stem cell sources. Moreover, we are beginning to realise the possibilities of using endogenous populations of cells from within the retina itself to mediate retinal repair. Here, we present a review of our current understanding of endogenous repair mechanisms together with recent progress in the use of both ocular and pluripotent stem cells for the treatment of photoreceptor loss. -

Repressing Notch Signaling and Expressing TNF Are Sufficient To
The Journal of Neuroscience, October 22, 2014 • 34(43):14403–14419 • 14403 Development/Plasticity/Repair Repressing Notch Signaling and Expressing TNF␣ Are Sufficient to Mimic Retinal Regeneration by Inducing Mu¨ller Glial Proliferation to Generate Committed Progenitor Cells Clay Conner, Kristin M. Ackerman, Manuela Lahne, Joshua S. Hobgood, and David R. Hyde Department of Biological Sciences and the Center for Zebrafish Research, Galvin Life Sciences Building, University of Notre Dame, Notre Dame, Indiana 46556 Retinal damage in teleosts, unlike mammals, induces robust Mu¨ller glia-mediated regeneration of lost neurons. We examined whether Notch signaling regulates Mu¨ller glia proliferation in the adult zebrafish retina and demonstrated that Notch signaling maintains Mu¨ller glia in a quiescent state in the undamaged retina. Repressing Notch signaling, through injection of the ␥-secretase inhibitor RO4929097, stimulates a subset of Mu¨ller glia to reenter the cell cycle without retinal damage. This RO4929097-induced Mu¨ller glia proliferation is mediated by repressing Notch signaling because inducible expression of the Notch Intracellular Domain (NICD) can reverse the effect. ThisRO4929097-inducedproliferationrequiresAscl1aexpressionandJak1-mediatedStat3phosphorylation/activation,analogoustothe light-damagedretina.Moreover,coinjectingRO4929097andTNF␣,apreviouslyidentifieddamagesignal,inducedthemajorityofMu¨ller glia to reenter the cell cycle and produced proliferating neuronal progenitor cells that committed to a neuronal lineage in