A Signature of a Recent Selective Sweep Identifies a Mutation That Defines the Cornish Rex Cat Breed." Plos One.8,6
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Prepubertal Gonadectomy in Male Cats: a Retrospective Internet-Based Survey on the Safety of Castration at a Young Age
ESTONIAN UNIVERSITY OF LIFE SCIENCES Institute of Veterinary Medicine and Animal Sciences Hedvig Liblikas PREPUBERTAL GONADECTOMY IN MALE CATS: A RETROSPECTIVE INTERNET-BASED SURVEY ON THE SAFETY OF CASTRATION AT A YOUNG AGE PREPUBERTAALNE GONADEKTOOMIA ISASTEL KASSIDEL: RETROSPEKTIIVNE INTERNETIKÜSITLUSEL PÕHINEV NOORTE KASSIDE KASTREERIMISE OHUTUSE UURING Graduation Thesis in Veterinary Medicine The Curriculum of Veterinary Medicine Supervisors: Tiia Ariko, MSc Kaisa Savolainen, MSc Tartu 2020 ABSTRACT Estonian University of Life Sciences Abstract of Final Thesis Fr. R. Kreutzwaldi 1, Tartu 51006 Author: Hedvig Liblikas Specialty: Veterinary Medicine Title: Prepubertal gonadectomy in male cats: a retrospective internet-based survey on the safety of castration at a young age Pages: 49 Figures: 0 Tables: 6 Appendixes: 2 Department / Chair: Chair of Veterinary Clinical Medicine Field of research and (CERC S) code: 3. Health, 3.2. Veterinary Medicine B750 Veterinary medicine, surgery, physiology, pathology, clinical studies Supervisors: Tiia Ariko, Kaisa Savolainen Place and date: Tartu 2020 Prepubertal gonadectomy (PPG) of kittens is proven to be a suitable method for feral cat population control, removal of unwanted sexual behaviour like spraying and aggression and for avoidance of unwanted litters. There are several concerns on the possible negative effects on PPG including anaesthesia, surgery and complications. The aim of this study was to evaluate the safety of PPG. Microsoft excel was used for statistical analysis. The information about 6646 purebred kittens who had gone through PPG before 27 weeks of age was obtained from the online retrospective survey. Database included cats from the different breeds and –age groups when the surgery was performed, collected in 2019. -

New Zealand Cat Fancy Inc. SHORTHAIR DIVISION Standards
Issued 2017 Member of the World Cat Congress New Zealand Cat Fancy Inc. SHORTHAIR DIVISION Standards of Points Issued 2017 Amendment Summary 31 Jan 2017 Introduction Updated Unable to be Judged to replace UTJ Cat 2 with Unable to be Handled, and removed Section 3 categories. (Ref: 17-006.) BUR Colour Charts: Updates to russet introduction and descriptions. (Ref: 17-015.) Reformatted with minor changes not affecting content. MDY Colour Charts: Updates to russet introduction and descriptions. (Ref: 17-016.) Reformatted with minor changes not affecting content. BEN General Type Standard and Scale of Points: Significant changes to align better with TICA standard. (Ref: 17-018, 17-020.) CAS General Type Standard and Scale of Points: Significant changes to align better with TICA standard. (Ref: 17-019, 17-021.) 12 Apr 2016 Introduction Removed intentionally blank page and heading pages for Parts 1 and 2. (Ref: 16-023.) General SIA, BAL, JAV, Added blank pages to assist with double-sided printing. Note: Issue dates not changed. TMA / TCM, (Ref: 16-025.) TRS / TRL, ABY, TIF, BML / BLH, RUS, TYG, AUM Amendment Process 0.1 Suggestions for minor amendments (minor errors or omissions which do not affect the intent) are welcome and may be submitted to the Secretary. These will usually be incorporated the next time the standard is reissued. 0.2 Proposals for significant amendments should also be submitted to the Secretary but will require a process of assessment, consultation, and approval prior to changes being made. INTRO-2 NZCF SH Standard of Points – Introduction Issued 2017 Contents 1. Show Groups - Breed Codes .................................................................................................. -

2019 Proceedings Book
2019 32ND PROCEEDINGS OF April 10-13 HILTON AUSTIN NEW ORLEANS APRIL 21-24 SHERATON NEW ORLEANS HOTEL 2 Sydney is closer than you think. Follow us for updates vetdermsydney.com Principal Sponsors Major Sponsors 3 TABLE OF CONTENTS GENERAL INFORMATION ABSTRACTS #detectDex 5 THURSDAY 19 App 5 Resident Abstract Presentations 21 Hotel Map 6 ISVD Sessions 45 Registration Hours 7 Concurrent Session Presentations 63 Exhibit Hall Hours 7 Poster Hours 7 FRIDAY 78 Exhibit Hall Map 7 Original Abstract Presentations 80 Sponsors 8 Clinical Abstract Presentations 99 Exhibitors 9 Scientific Session Presentations 103 Concurrent Session Presentations 105 COMPLETE SCHEDULE Wednesday 10 SATURDAY 118 Thursday 11 Clinical Abstract Presentations 120 Friday 14 Scientific Session Presentations 131 Saturday 16 Concurrent Session Presentations 158 ADVT Sessions 179 ROUNDTABLE SESSIONS POSTERS 184 Thursday 18 Friday 18 Saturday 18 4 Help us Keep Track of Dex! Dex is ready to explore Austin. While we’d love for him to sample some BBQ and jam out on South Sixth Street, we want to make sure he’s not getting into any trouble. Help us keep track of him during the conference. If you spot him make sure to snap a photo and share it on the app using your Instagram account. Remember to tag NAVDF (@navdf) and use the hashtags #detectDex, #NAVDF, and #NAVDF2019. Once your photo is shared, return Dex to his dog house at NAVDF registration and claim your reward! ® APP DOWNLOAD INSTRUCTIONS 1. Search NAVDF in the iTunes or Google Play Store. 2. Tap “Get” or “Install” OR LAPTOP OR OTHER DEVICES Enter https://crowd.cc/2xzru in your browser search bar 5 HOTEL MEETING SPACE 6 REVISION Date:2/7/2019 REGISTRATIONAMERICAN & EXHIBIT ACADEMY OF HALL VETERINARY HOURSBy: MAREESA JOHNSON DERMATOLOGY BOOTH COUNT APRIL 11-13, 2019 Inventory as of 02/07/2019 Dimension Size Qty SqFt 8'x10' 80 49 3,920 HILTON AUSTIN DOWNTOWN - GRAND BALLROOM SALON H - AUSTIN,TX Totals: 49 3,920 REGISTRATION INFORMATION EXHIBIT HALL & POSTERBLDG. -

Clinical and Histologic Description of Lykoi Cat Hair Coat and Skin
獣医臨床皮膚科 22 (3): 179–191, 2016 Original Clinical and Histologic Description of Lykoi Cat Hair Coat and Skin リコイ猫の被毛と皮膚に関する臨床的および組織学的記述 Michelle L. LeRoy1, 2)*, David A. Senter1, 2), Dae Young Kim3), Barbara Gandolfi2), John R. Middleton2), Karen E. Trainor4), Delia M. Bouhan2), Leslie A. Lyons2) 1)Veterinary Allergy and Dermatology Clinic, LLC, 2)Department of Veterinary Medicine and Surgery, University of Missouri, College of Veterinary Medicine, 3)Department of Veterinary Pathobiology, University of Missouri, College of Veterinary Medicine, 4)Innovative Vet Path, LLC Received April 9, 2016 and accepted June 7, 2016 Abstract: Hair and skin abnormalities of domesticated animals are readily identified and are biomedical models for ectodermal dysplasias. The hair coat of the Lykoi cat, a new cat breed, is a dramatic phenotype and has not been clinically or histologically described. Dermatoscopic examination was performed and skin biopsies were collected from seven Lykoi cats and seven dermatologically normal domestic shorthair (DSH) cats. All skin structures were examined on longitudinal and transverse sections. Immunohistochemistry for CD3 and Cytokeratin 8/18 was performed for comparison with DSH cats. Dermatoscopic images were compared. Lykoi had a significant reduction in average numbers of follicles per hair follicle group as compared to DSH cats, 14.7 ± 2.9 and 23.4 ± 5.4, respectively. Median (range) numbers of hairs per hair follicle group were 1.3 (0.4–5.7) and 18.8 (10.6–26.6), respectively. Mean (± SD) hair follicle depth was 0.95 mm ± 0.15 and 1.14 mm ± 0.21 for Lykoi and DSH cats, respectively. Mean (± SD) primary hair shaft diameters were 39 µm ± 0.029 and 47 µm ± 0.011 for Lykoi and DSH cats, respectively. -

Liste Der Rassen, Welche Von Der WCF Anerkannt Und Akzeptiert Sind
List of breeds, which are recognized and admitted by the WCF Breed Synonym Category Recognized Admitted Preliminary WCC-Organization LOOF Abyssinian SH x all American SH x CFA, TICA, Bobtail SH LOOF American SLH x CFA, TICA, Bobtail LH LOOF American Curl SH x CFA, FIFe, SACC, SH TICA, LOOF American Curl SLH x CFA, FIFe, SACC, LH TICA, LOOF American SH x CFA, SACC, TICA, Shorthair LOOF American SH x CFA, TICA, Wirehair LOOF Anatoli SH x Arabian Mau SH x x Asian SH x CCCA:Self, GCCF, SACC, LOOF Australian SH x ACF, CCCA, NZCF Mist Balinese SLH x all Bengal SH x ACF, CCCA, FIFe, GCCF, NZCF, SACC, TICA, LOOF Bombay Asian SH x ACF, CCCA, CFA, GCCF: Asian, NZCF, SACC: Asian, TICA, LOOF Brazilian SH x Shorthair British SH x all Shorthair British SLH x TICA: new trait class, Longhair LOOF Burmese 1) SH x all Burmilla Asian SH x ACF, CCCA, FIFe, GCCF: Asian, NZCF, SACC: Asian, LOOF: Asian Burmilla LH SLH x ACF, CCCA Celtic European SH SH x ACF, FIFe, Shorthair LOOF Ceylon SH x LOOF Chartreux SH x ACF, CFA, FIFe, TICA, Edition 01-04-2009 1/5 List of breeds, which are recognized and admitted by the WCF LOOF Chausie SH x x TICA, LOOF Classicat Ocicat blotched SH x NZCF Colourpoint Himalayan LH x all Colourpoint Siamese: all other SOSH x all Shorthair colours than the 4 basic colours Cornish Rex SH x all Cymric Manx LH SLH x ACF, CCCA, CFA, FIFe, NZCF, SACC, TICA, LOOF Devon Rex SH x all Don Sphynx Donskoy SH x FIFe, TICA: preliminary, LOOF Egyptian Mau SH x ACF, CCCA, CFA, FIFe, GCCF, NZCF, TICA, LOOF Exotic Exotic SH x all Shorthair Foreign White -

R for Rexcellence
Petcare Hints R for Rexcellence Rex cats make excellent pets. Their curly-coated charisma invariably charms all those exposed to their impish ways. These are fascinating felines with an offbeat style all their own. Over the years there has been a variety of Rex Cats evolve around the world, including the German Rex, Oregon Rex, Ohio Rex and more recently the Selkirk Rex. However it is the Cornish Rex and the Devon Rex which have gained by far the greatest recognition and popularity. Although tales of curly-coated cats circulated throughout the west of England for many years, it was not until 1950 that the first one was officially documented. Two farm cats living near Bodmin Moor in Cornwall produced a curly-coated kitten (subsequently called Kallibunker) from an otherwise unremarkable litter. Fortunately the owner had previously bred and shown Astrex rabbits (which have the same characteristic Rex-coat type) so she understood the significance of this spontaneous genetic mutation. This heralded the start of the Cornish Rex breed. Ten years later, in the adjoining county, another curly-coated kitten appeared in a litter produced by two domestic shorthaired cats. Again, although coming from ordinary "moggies", the kitten in question (called Kirlee) looked like it was of a foreign breed. Naturally, this cat from Devon was assumed to be the same as that originating in Cornwall, but when Kirlee was mated to Kallibunker's descendants, on each occasion only straight-coated kittens resulted. It was subsequently realised that the Cornish Rex and Devon Rex each possessed different genes for curly coats - and were in fact two separate "breeds". -

National Breed Standards
NATIONAL BREED STANDARDS © ANCATS 2017 ANCATS National Breed Standards 2017 1 INDEX ........................................................................................................................................................................................................................................................................... 2 GLOSSARY ................................................................................................................................................................................................................................................................. 5 PREFACE .................................................................................................................................................................................................................................................................. 10 The Condition of the Cat .................................................................................................................................................................................................................. 11 Judging Disqualification Faults ........................................................................................................................................................................................................ 12 General faults in all breeds precluding a Challenge or Best in Show ............................................................................................................................................. -

Breeding Rules
THIS TRANSLATION SERVES ONLY FOR EASY UNDERSTANDING. IN CASE OF DOUBTS OR ERRORS ONLY THE GERMAN VERSION IS VALID. Healthrules Husbandryrules Breedingrules 1.7.2021 © KKÖ Health-/Husbandry- and Breeding-rules The breeding-rules of the KKÖ are based on the currently valid rules of the FIFe and the currently valid version of the Animal Protection act. 1. REQUIREMENTS AND NECESSARY STEPS IN BREEDING In the following, the requirements, what you need and should consider for breeding, as well as the necessary steps that you must take, are explained. Before you can start breeding and call yourself a breeder, you must be an unterminated ordinary member of the KKÖ and have paid your membership fee. What breeding means and who is a breeder is in Art. 2-a. and 2-b. explained. The first step is to apply for a cattery name. The rule for cattery names is in Art. 2-c. explained. You must be the owner of a female cat who follows the rules in Art. 3. What owner of a female breeding cat means is in Art. 2-b. explained. For breeding you need a stud male. The male cat, the own or a third party one, must also correspond to certain regulations which are explained Art. 4. If you want to buy a female or male cat, please follow the rules in Art. 7-c.5. up to 7- c.7. For the mating of a female and a male cat please follow the rules in Art 5. After the birth of the kittens, the announcement must be submitted to the breeding committee of the KKÖ. -

REX the KING the Rex Cats, So New to the Fancy That Many Fanciers Who
REX THE KING By Helen Weiss The Rex cats, so new to the fancy that many Fanciers who show, and even some of the judges have never seen one, are of the most out standing breed. Most new breeds and colors are man made by hybridiz ing between two or more breeds and work ing with them until the desired character istics are obtained. This is not true with the Rex for it is a mutation from the Do mestic Shorthair and can be obtained only when both parents carry the rex gene or by the original mutation created by God. Many think that a Rex cat is just cat without guardhairs. This is not true for these cats, even in the first mutation, are as unlike their normal coated siblings as they can be. Almost without fail these cats show a definitely foreign type. The differ ence is best seen in the illustration of the Rex cat from Ohio and his litter brother. The Rex are outstanding in appear ance, their most distinguishing characteristic being' their short, dense and usually curly coat that is so soft. to the touch that one can never forget the feel of this luxurious fur. This fur is different from that of the normal cat, for it has no g'uard hairs, with the und~r coat or down hairs shorter and finer but more numerous than the hall' on any other breed. 1 Rex cats have a delicate appearance which belies their heavy weight and muscular body. Most Rex cats are fine boned and slim in appearance, but part of this is an illusion created by the very short coat, so that one is able to see the muscle forms that are covered by a longer coat in other cats. -
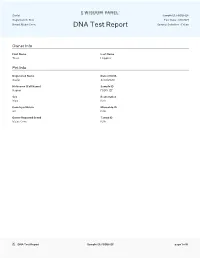
Davlat's DNA Test Report
Davlat Sample ID: FBDSHZF Registration: N/A Test Date: 2/9/2021 Breed: Maine Coon DNA Test Report Optimal Selection - Feline Owner Info First Name Last Name Traci Heppner Pet Info Registered Name Date of Birth Davlat 3/24/2020 Nickname (Call Name) Sample ID Ragnar FBDSHZF Sex Registration Male N/A Country of Origin Microchip ID RU N/A Owner Reported Breed Tattoo ID Maine Coon N/A DNA Test Report Sample ID: FBDSHZF page 1 of 6 Davlat Sample ID: FBDSHZF Registration: N/A Test Date: 2/9/2021 Breed: Maine Coon DNA Test Report Optimal Selection - Feline Health Conditions Known in This Breed Genetic Condition Gene Risk Variant Copies Result Cystinuria Type B (Variant 3) SCL7A9 T>A 0 Clear Factor XII Deficiency (Variant 1) F12 Deletion 0 Clear Factor XII Deficiency (Variant 2) F12 Deletion 0 Clear Hypertrophic Cardiomyopathy (A31P; Discovered in Maine Coon) MYBPC G>C 0 Clear Pyruvate Kinase Deficiency PLKR G>A 0 Clear Spinal Muscular Atrophy (Discovered in Maine Coon) LIX1 Deletion 0 Clear Other Conditions Tested Genetic Condition Gene Risk Variant Copies Result Acute Intermittent Porphyria (Variant 1) AIP Deletion 0 Clear Acute Intermittent Porphyria (Variant 2) AIP G>A 0 Clear Acute Intermittent Porphyria (Variant 3) HMBS Insertion 0 Clear Acute Intermittent Porphyria (Variant 4) HMBS Deletion 0 Clear Acute Intermittent Porphyria (Variant 5) HMBS G>A 0 Clear Autoimmune Lymphoproliferative Syndrome FASL Insertion 0 Clear Burmese Head Defect (Discovered in Burmese) ALX1 Deletion 0 Clear Chediak-Higashi Syndrome (Discovered in Persian cats) CHS -

LOOF STANDARDS Table of Contents
LOOF STANDARDS Table of contents ABYSSINIAN & S OMALI MAINE C OON AMERICAN B OBTAIL S HORTHAIR & L ONGHAIR MANX & C YMRIC AMERICAN C URL S HORTHAIR & L ONGHAIR MUNCHKIN S HORTHAIR & L ONGHAIR AMERICAN S HORTHAIR NORWEGIAN F OREST C AT AMERICAN W IREHAIR OCICAT ASIAN , E NGLISH B URMESE & B URMILLA PERSIAN & E XOTIC S HORTHAIR BENGAL PETERBALD BIRMAN PIXIE B OB S HORTHAIR & L ONGHAIR BOMBAY & A MERICAN B URMESE RAGDOLL BRITISH S HORTHAIR & L ONGHAIR RUSSIAN & N EBELUNG CALIFORNIA S PANGLED C AT SAVANNAH CEYLON SCOTTISH & H IGHLAND CHARTREUX SELKIRK CHAUSIE SIAMESE , O RIENTAL , B ALINESE & M ANDARIN CORNISH R EX & C ALIFORNIAN R EX SIBERIAN DEVON R EX SINGAPURA DONSKOY SNOWSHOE EUROPEAN S HORTHAIR SOKOKE EGYPTIAN M AU SPHYNX GERMAN R EX THAI HAVANA B ROWN TONKINESE S HORTHAIR & L ONGHAIR JAPANESE B OBTAIL S HORTHAIR & L ONGHAIR TURKISH A NGORA KORAT TURKISH V AN KURILIAN B OBTAIL S HORTHAIR & L ONGHAIR YORK C HOCOLATE LAPERM S HORTHAIR & L ONGHAIR LOOF standards TABLE OF CONTENTS English translation of version revised 29 November 2010 LOOF standards TABLE OF CONTENTS English translation of version revised 29 November 2010 ABYSSINIAN & SOMALI HEAD = 30 points royal look. Medium in size, males are Profile = 5 proportionally larger than females. Muzzle = 5 Well muscled, the Abyssinian and the Skull = 5 Somali are supple and agile like Ears = 5 panthers and show a great interest in Eye shape = 5 their environment. Their ticked coat Neck = 5 has the quality to reflect the light. Because of his hair length, the Somali BODY = 35 points may seem a bit heavier than he really Torso = 10 is. -

Using Petlink's Mass Import Feature
Using PetLink’s Mass Import Feature Instructions for Animal Professionals (Shelters, Rescues and Humane Societies) Overview PetLink offers Mass Import of multiple prepaid microchip registrations and transfers of ownership for Animal Professionals, so that you can upload pet data in bulk, saving valuable time entering individual records. PetLink’s Mass Import is based on a simple, easy to use Microsoft Excel spreadsheet template, where owner and pet data are input into one document, which then can be uploaded into PetLink directly at one time. Mass Import enables immediate registration, and eliminates the need for individual registration and transfer of ownership forms that have to be mailed or faxed to PetLink. Mass Import also helps ensure that pet owner contact data Guardian: remains in secure hands. We are committed to our customers’ Linking You, Your Clients and Pets privacy; Mass Import creates an efficient and effective means to complete registration or transfer of ownership quickly By using PetLink’s Mass Import feature, your facility will and ensures that the privacy of both you and your clients is automatically be listed as “guardian” for every pet registered - protected. Your facility will receive a confirmation report and something that a pet owner or adopter cannot edit. This adds your each individual pet owner will receive a PetLink welcome email facility as a permanent contact in case the pet is ever found far following a successful import. from home but its family cannot be reached. It helps ensure that you can remain involved in the pet’s care and in making decisions In order to use Mass Import, your facility needs to have a valid, on how it is to be treated and possibly rehomed if that animal is free, Animal Professional account with PetLink.