Fourier Transform Ion Cyclotron Resonance Mass Spectrometer in Metabolomics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Metabolic Engineering of Microbial Cell Factories for Biosynthesis of Flavonoids: a Review
molecules Review Metabolic Engineering of Microbial Cell Factories for Biosynthesis of Flavonoids: A Review Hanghang Lou 1,†, Lifei Hu 2,†, Hongyun Lu 1, Tianyu Wei 1 and Qihe Chen 1,* 1 Department of Food Science and Nutrition, Zhejiang University, Hangzhou 310058, China; [email protected] (H.L.); [email protected] (H.L.); [email protected] (T.W.) 2 Hubei Key Lab of Quality and Safety of Traditional Chinese Medicine & Health Food, Huangshi 435100, China; [email protected] * Correspondence: [email protected]; Tel.: +86-0571-8698-4316 † These authors are equally to this manuscript. Abstract: Flavonoids belong to a class of plant secondary metabolites that have a polyphenol structure. Flavonoids show extensive biological activity, such as antioxidative, anti-inflammatory, anti-mutagenic, anti-cancer, and antibacterial properties, so they are widely used in the food, phar- maceutical, and nutraceutical industries. However, traditional sources of flavonoids are no longer sufficient to meet current demands. In recent years, with the clarification of the biosynthetic pathway of flavonoids and the development of synthetic biology, it has become possible to use synthetic metabolic engineering methods with microorganisms as hosts to produce flavonoids. This article mainly reviews the biosynthetic pathways of flavonoids and the development of microbial expression systems for the production of flavonoids in order to provide a useful reference for further research on synthetic metabolic engineering of flavonoids. Meanwhile, the application of co-culture systems in the biosynthesis of flavonoids is emphasized in this review. Citation: Lou, H.; Hu, L.; Lu, H.; Wei, Keywords: flavonoids; metabolic engineering; co-culture system; biosynthesis; microbial cell factories T.; Chen, Q. -

Key Enzymes Involved in the Synthesis of Hops Phytochemical Compounds: from Structure, Functions to Applications
International Journal of Molecular Sciences Review Key Enzymes Involved in the Synthesis of Hops Phytochemical Compounds: From Structure, Functions to Applications Kai Hong , Limin Wang, Agbaka Johnpaul , Chenyan Lv * and Changwei Ma * College of Food Science and Nutritional Engineering, China Agricultural University, 17 Qinghua Donglu Road, Haidian District, Beijing 100083, China; [email protected] (K.H.); [email protected] (L.W.); [email protected] (A.J.) * Correspondence: [email protected] (C.L.); [email protected] (C.M.); Tel./Fax: +86-10-62737643 (C.M.) Abstract: Humulus lupulus L. is an essential source of aroma compounds, hop bitter acids, and xanthohumol derivatives mainly exploited as flavourings in beer brewing and with demonstrated potential for the treatment of certain diseases. To acquire a comprehensive understanding of the biosynthesis of these compounds, the primary enzymes involved in the three major pathways of hops’ phytochemical composition are herein critically summarized. Hops’ phytochemical components impart bitterness, aroma, and antioxidant activity to beers. The biosynthesis pathways have been extensively studied and enzymes play essential roles in the processes. Here, we introduced the enzymes involved in the biosynthesis of hop bitter acids, monoterpenes and xanthohumol deriva- tives, including the branched-chain aminotransferase (BCAT), branched-chain keto-acid dehydroge- nase (BCKDH), carboxyl CoA ligase (CCL), valerophenone synthase (VPS), prenyltransferase (PT), 1-deoxyxylulose-5-phosphate synthase (DXS), 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (HDR), Geranyl diphosphate synthase (GPPS), monoterpene synthase enzymes (MTS), cinnamate Citation: Hong, K.; Wang, L.; 4-hydroxylase (C4H), chalcone synthase (CHS_H1), chalcone isomerase (CHI)-like proteins (CHIL), Johnpaul, A.; Lv, C.; Ma, C. -

Identification of Compounds with Potential Therapeutic Uses From
International Journal of Molecular Sciences Article Identification of Compounds with Potential Therapeutic Uses from Sweet Pepper (Capsicum annuum L.) Fruits and Their Modulation by Nitric Oxide (NO) Lucía Guevara 1, María Ángeles Domínguez-Anaya 1, Alba Ortigosa 1, Salvador González-Gordo 1 , Caridad Díaz 2 , Francisca Vicente 2 , Francisco J. Corpas 1 , José Pérez del Palacio 2 and José M. Palma 1,* 1 Group of Antioxidant, Free Radicals and Nitric Oxide in Biotechnology, Food and Agriculture, Department of Biochemistry, Cell and Molecular Biology of Plants, Estación Experimental del Zaidín, CSIC, 18008 Granada, Spain; [email protected] (L.G.); [email protected] (M.Á.D.-A.); [email protected] (A.O.); [email protected] (S.G.-G.); [email protected] (F.J.C.) 2 Department of Screening & Target Validation, Fundación MEDINA, 18016 Granada, Spain; [email protected] (C.D.); [email protected] (F.V.); [email protected] (J.P.d.P.) * Correspondence: [email protected]; Tel.: +34-958-181-1600; Fax: +34-958-181-609 Abstract: Plant species are precursors of a wide variety of secondary metabolites that, besides being useful for themselves, can also be used by humans for their consumption and economic benefit. Pepper (Capsicum annuum L.) fruit is not only a common food and spice source, it also stands out for containing high amounts of antioxidants (such as vitamins C and A), polyphenols and capsaicinoids. Citation: Guevara, L.; Particular attention has been paid to capsaicin, whose anti-inflammatory, antiproliferative and Domínguez-Anaya, M.Á.; Ortigosa, A.; González-Gordo, S.; Díaz, C.; analgesic activities have been reported in the literature. -

“Mechanical Universe and Beyond” Videos—CE Mungan, Spring 2001
Keywords in “Mechanical Universe and Beyond” Videos—C.E. Mungan, Spring 2001 This entire series can be viewed online at http://www.learner.org/resources/series42.html. Some of the titles below have been modified by me to better reflect their contents. In my opinion, tapes 21–22 are the best in the whole series! 1. Introduction to Classical Mechanics: Kepler, Galileo, Newton 2. Falling Bodies: s = gt2 / 2, ! = gt, a = g 3. Differentiation: introductory math 4. Inertia: Newton’s first law, Copernican solar system 5. Vectors: quaternions, unit vectors, dot and cross products 6. Newton’s Laws: Newton’s second law, momentum, Newton’s third law, monkey-gun demo 7. Integration: Newton vs. Leibniz, anti-derivatives 8. Gravity: planetary orbits, universal law of gravity, the Moon falls toward the Earth 9. UCM: Ptolemaic solar system, centripetal acceleration and force 10. Fundamental Forces: Cavendish experiment, Franklin, unified theory, viscosity, tandem accelerator 11. Gravity and E&M: fundamental constants, speed of light, Oersted experiment, Maxwell 12. Millikan Experiment: CRT, scientific method 13. Energy Conservation: work, gravitational PE, KE, mechanical energy, heat, Joule, microscopic forms of energy, useful available energy 14. PE: stability, conservation, position dependence, escape speed 15. Conservation of Linear Momentum: Descartes, generalized Newton’s second law, Earth- Moon system, linear accelerator 16. SHM: amplitude-independent period of pendulum, timekeeping, restoring force, connection to UCM, elastic PE 17. Resonance: Tacoma Narrows, music, breaking wineglass demo, earthquakes, Aeolian harp, vortex shedding 18. Waves: shock waves, speed of sound, coupled oscillators, wave properties, gravity waves, isothermal vs. adiabatic bulk modulus 19. Conservation of Angular Momentum: Kepler’s second law, vortices, torque, Brahe 20. -

L.) Leaves Tao Jiang1,3, Kunyuan Guo2,3, Lingdi Liu1, Wei Tian1, Xiaoliang Xie1, Saiqun Wen1 & Chunxiu Wen1*
www.nature.com/scientificreports OPEN Integrated transcriptomic and metabolomic data reveal the favonoid biosynthesis metabolic pathway in Perilla frutescens (L.) leaves Tao Jiang1,3, Kunyuan Guo2,3, Lingdi Liu1, Wei Tian1, Xiaoliang Xie1, Saiqun Wen1 & Chunxiu Wen1* Perilla frutescens (L.) is an important medicinal and edible plant in China with nutritional and medical uses. The extract from leaves of Perilla frutescens contains favonoids and volatile oils, which are mainly used in traditional Chinese medicine. In this study, we analyzed the transcriptomic and metabolomic data of the leaves of two Perilla frutescens varieties: JIZI 1 and JIZI 2. A total of 9277 diferentially expressed genes and 223 favonoid metabolites were identifed in these varieties. Chrysoeriol, apigenin, malvidin, cyanidin, kaempferol, and their derivatives were abundant in the leaves of Perilla frutescens, which were more than 70% of total favonoid contents. A total of 77 unigenes encoding 15 enzymes were identifed as candidate genes involved in favonoid biosynthesis in the leaves of Perilla frutescens. High expression of the CHS gene enhances the accumulation of favonoids in the leaves of Perilla frutescens. Our results provide valuable information on the favonoid metabolites and candidate genes involved in the favonoid biosynthesis pathways in the leaves of Perilla frutescens. Perilla frutescens (L.), which is a self-compatible annual herb, belongs to the family Lamiaceae. Tis species has been widely cultivated in China, Japan, and Korea for centuries. Perilla frutescens is an important medicinal and edible plant in China with medical and nutritional uses 1. Its leaves can be utilized as a transitional medicinal herb, as a vegetable, and as a spice, and its seeds can be processed into foods and nutritional edible oils 2. -

Caltech News
Volume 16, No.7, December 1982 CALTECH NEWS pounds, became optional and were Three Caltech offered in the winter and spring. graduate programs But under this plan, there was an overlap in material that diluted the rank number one program's efficiency, blending per in nationwide survey sons in the same classrooms whose backgrounds varied widely. Some Caltech ranked number one - students took 3B and 3C before either alone or with other institutions proceeding on to 46A and 46B, - in a recent report that judged the which focused on organic systems, scholastic quality of graduate pro" while other students went directly grams in mathematics and science at into the organic program. the nation's major research Another matter to be addressed universities. stemmed from the fact that, across Caltech led the field in geoscience, the country, the lines between inor and shared top rankings with Har ganic and organic chemistry had ' vard in physics. The Institute was in become increasingly blurred. Explains a four-way tie for first in chemistry Professor of Chemistry Peter Der with Berkeley, Harvard, and MIT. van, "We use common analytical The report was the result of a equipment. We are both molecule two-year, $500,000 study published builders in our efforts to invent new under the sponsorship of four aca materials. We use common bonds for demic groups - the American Coun The Mead Laboratory is the setting for Chemistry 5, where Carlotta Paulsen uses a rotary probing how chemical bonds are evaporator to remove a solvent from a synthesized product. Paulsen is a junior majoring in made and broken." cil of Learned Societies, the American chemistry. -

Nuclear Magnetic Resonance and Its Application in Condensed Matter Physics
Nuclear Magnetic Resonance and Its Application in Condensed Matter Physics Kangbo Hao 1. Introduction Nuclear Magnetic Resonance (NMR) is a physics phenomenon first observed by Isidor Rabi in 1938. [1] Since then, the NMR spectroscopy has been applied in a wide range of areas such as physics, chemistry, and medical examination. In this paper, I want to briefly discuss about the theory of NMR spectroscopy and its recent application in condensed matter physics. 2. Principles of NMR NMR occurs when some certain nuclei are in a static magnetic field and another oscillation magnetic field. Assuming a nucleus has a spin angular momentum 퐼⃗ = ℏ푚퐼, then its magnetic moment 휇⃗ is 휇⃗ = 훾퐼⃗ (1) The 훾 here is the gyromagnetic ratio, which depends on the property of the nucleus. If we put such a nucleus in a static magnetic field 퐵⃗⃗0, then the magnetic moment of this nuclei will process about this magnetic field. Therefore we have, [2] [3] 푑퐼⃗ 1 푑휇⃗⃗⃗ 휏⃗ = 휇⃗ × 퐵⃗⃗ = = (2) 0 푑푥 훾 푑푥 From this semiclassical picture, we can easily derive that the precession frequency 휔0 (which is called the Larmor angular frequency) is 휔0 = 훾퐵0 (3) Then, if another small oscillating magnetic field is added to the plane perpendicular to 퐵⃗⃗0, then the total magnetic field is (Assuming 퐵⃗⃗0 is in 푧̂ direction) 퐵⃗⃗ = 퐵0푧̂ + 퐵1(cos(휔푡) 푥̂ + sin(휔푡) 푦̂) (4) If we choose a frame (푥̂′, 푦̂′, 푧̂′ = 푧̂) rotating with the oscillating magnetic field, then the effective magnetic field in this frame is 휔 퐵̂ = (퐵 − ) 푧̂ + 퐵 푥̂′ (5) 푒푓푓 0 훾 1 As a result, at 휔 = 훾퐵0, which is the resonant frequency, the 푧̂ component will vanish, and thus the spin angular momentum will precess about 퐵⃗⃗1 instead. -

Chemistry and Pharmacology of Kinkéliba (Combretum
CHEMISTRY AND PHARMACOLOGY OF KINKÉLIBA (COMBRETUM MICRANTHUM), A WEST AFRICAN MEDICINAL PLANT By CARA RENAE WELCH A Dissertation submitted to the Graduate School-New Brunswick Rutgers, The State University of New Jersey in partial fulfillment of the requirements for the degree of Doctor of Philosophy Graduate Program in Medicinal Chemistry written under the direction of Dr. James E. Simon and approved by ______________________________ ______________________________ ______________________________ ______________________________ New Brunswick, New Jersey January, 2010 ABSTRACT OF THE DISSERTATION Chemistry and Pharmacology of Kinkéliba (Combretum micranthum), a West African Medicinal Plant by CARA RENAE WELCH Dissertation Director: James E. Simon Kinkéliba (Combretum micranthum, Fam. Combretaceae) is an undomesticated shrub species of western Africa and is one of the most popular traditional bush teas of Senegal. The herbal beverage is traditionally used for weight loss, digestion, as a diuretic and mild antibiotic, and to relieve pain. The fresh leaves are used to treat malarial fever. Leaf extracts, the most biologically active plant tissue relative to stem, bark and roots, were screened for antioxidant capacity, measuring the removal of a radical by UV/VIS spectrophotometry, anti-inflammatory activity, measuring inducible nitric oxide synthase (iNOS) in RAW 264.7 macrophage cells, and glucose-lowering activity, measuring phosphoenolpyruvate carboxykinase (PEPCK) mRNA expression in an H4IIE rat hepatoma cell line. Radical oxygen scavenging activity, or antioxidant capacity, was utilized for initially directing the fractionation; highlighted subfractions and isolated compounds were subsequently tested for anti-inflammatory and glucose-lowering activities. The ethyl acetate and n-butanol fractions of the crude leaf extract were fractionated leading to the isolation and identification of a number of polyphenolic ii compounds. -

Metabolomics by UHPLC-Q-TOF Reveals Host Tree-Dependent Phytochemical Variation in Viscum Album L
plants Article Metabolomics by UHPLC-Q-TOF Reveals Host Tree-Dependent Phytochemical Variation in Viscum album L. Tim Jäger 1,2,3,†, Carla Holandino 1,4,* , Michelle Nonato de Oliveira Melo 4,5 , Evelyn Maribel Condori Peñaloza 4,5, Adriana Passos Oliveira 4, Rafael Garrett 5 , Gaétan Glauser 6 , Mirio Grazi 1, Hartmut Ramm 1, Konrad Urech 1 and Stephan Baumgartner 1,3,7,* 1 Society for Cancer Research, Hiscia Institute, Kirschweg 9, 4144 Arlesheim, Switzerland; [email protected] (T.J.); [email protected] (M.G.); [email protected] (H.R.); [email protected] (K.U.) 2 Center for Complementary Medicine, Institute for Infection Prevention and Hospital Epidemiology, Faculty of Medicine, University of Freiburg, Breisacher Str. 115b, 79106 Freiburg, Germany 3 Institute of Integrative Medicine, University of Witten/Herdecke, Gerhard-Kienle-Weg 4, 58313 Herdecke, Germany 4 Laboratório Multidisciplinar de Ciências Farmacêuticas, Pharmacy College, Federal University of Rio de Janeiro, Rio de Janeiro 21941-902, Brazil; [email protected] (M.N.d.O.M.); [email protected] (E.M.C.P.); [email protected] (A.P.O.) 5 Metabolomics Laboratory, Chemistry Institute, Federal University of Rio de Janeiro, Rio de Janeiro 21941-598, Brazil; [email protected] 6 Neuchatel Platform of Analytical Chemistry, University of Neuchâtel, Avenue de Bellevaux 51, 2000 Neuchâtel, Switzerland; [email protected] 7 Institute of Complementary and Integrative Medicine, University of Bern, Freiburgstrasse 46, 3010 Bern, Switzerland * Correspondence: [email protected] (C.H.); [email protected] (S.B.) † Deceased 1 March 2019. Citation: Jäger, T.; Holandino, C.; Abstract: Viscum album L., commonly known as European mistletoe, is a hemi-parasitic plant of Melo, M.N.d.O.; Peñaloza, E.M.C.; the Santalaceae family. -
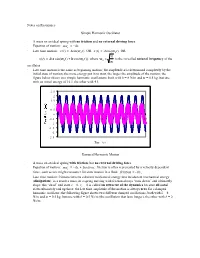
Notes on Resonance Simple Harmonic Oscillator • a Mass on An
Notes on Resonance Simple Harmonic Oscillator · A mass on an ideal spring with no friction and no external driving force · Equation of motion: max = - kx · Late time motion: x(t) = Asin(w0 t) OR x(t) = Acos(w0 t) OR k x(t) = A(a sin(w t) + b cos(w t)), where w = is the so-called natural frequency of the 0 0 0 m oscillator · Late time motion is the same as beginning motion; the amplitude A is determined completely by the initial state of motion; the more energy put in to start, the larger the amplitude of the motion; the figure below shows two simple harmonic oscillations, both with k = 8 N/m and m = 0.5 kg, but one with an initial energy of 16 J, the other with 4 J. 2.5 2 1.5 1 0.5 0 -0.5 0 5 10 15 20 -1 -1.5 -2 -2.5 Time (s) Damped Harmonic Motion · A mass on an ideal spring with friction, but no external driving force · Equation of motion: max = - kx + friction; friction is often represented by a velocity dependent force, such as one might encounter for slow motion in a fluid: friction = - bv x · Late time motion: friction converts coherent mechanical energy into incoherent mechanical energy (dissipation); as a result a mass on a spring moving with friction always “runs down” and ultimately stops; this “dead” end state x = 0, vx = 0 is called an attractor of the dynamics because all initial states ultimately end up there; the late time amplitude of the motion is always zero for a damped harmonic oscillator; the following figure shows two different damped oscillations, both with k = 8 N/m and m = 0.5 kg, but one with b = 0.5 Ns/m (the oscillation that lasts longer), the other with b = 2 Ns/m. -

Synthesis of Unnatural Alkaloid Scaffolds by Exploiting Plant Polyketide Synthase
Synthesis of unnatural alkaloid scaffolds by exploiting plant polyketide synthase Hiroyuki Moritaa,b, Makoto Yamashitaa, She-Po Shia,1, Toshiyuki Wakimotoa,b, Shin Kondoc, Ryohei Katoc, Shigetoshi Sugioc,2, Toshiyuki Kohnod,2, and Ikuro Abea,b,2 aGraduate School of Pharmaceutical Sciences, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan; bJapan Science and Technology Agency, Core Research of Evolutional Science and Technology, 5 Sanbancho, Chiyoda-ku, Tokyo 102-0075, Japan; cBiotechnology Laboratory, Mitsubishi Chemical Group Science and Technology Research Center Inc., 1000 Kamoshida, Aoba, Yokohama, Kanagawa 227-8502, Japan; and dMitsubishi Kagaku Institute of Life Sciences, 11 Minamiooya, Machida, Tokyo 194-8511, Japan Edited by Jerrold Meinwald, Cornell University, Ithaca, NY, and approved July 12, 2011 (received for review May 15, 2011) HsPKS1 from Huperzia serrata is a type III polyketide synthase (PKS) club moss Huperzia serrata (HsPKS1) (19) (Fig. S1), using precur- with remarkable substrate tolerance and catalytic potential. Here sor-directed and structure-based approaches. As previously re- we present the synthesis of unnatural unique polyketide–alkaloid ported, HsPKS1 is a unique type III PKS that exhibits unusually hybrid molecules by exploiting the enzyme reaction using precur- broad substrate specificity to produce various aromatic polyke- sor-directed and structure-based approaches. HsPKS1 produced tides. For example, HsPKS1, which normally catalyzes the sequen- novel pyridoisoindole (or benzopyridoisoindole) with the 6.5.6- tial condensations of 4-coumaroyl-CoA (1) with three molecules fused (or 6.6.5.6-fused) ring system by the condensation of 2-carba- of malonyl-CoA (2) to produce naringenin chalcone (3)(Fig.1A), moylbenzoyl-CoA (or 3-carbamoyl-2-naphthoyl-CoA), a synthetic also accepts bulky N-methylanthraniloyl-CoA (4)asastarterto nitrogen-containing nonphysiological starter substrate, with two produce 1,3-dihydroxy-N-methylacridone (5), after three conden- molecules of malonyl-CoA. -

Structure Assignment and H/D-Exchange Behavior of Several Glycosylated Polyphenols Andreas H
University of the Pacific Scholarly Commons College of the Pacific aF culty Articles All Faculty Scholarship 1-1-2014 Structure assignment and H/D-exchange behavior of several glycosylated polyphenols Andreas H. Franz University of the Pacific, [email protected] Ilona Serebnitskaya University of the Pacific Gurbir Gudial University of the Pacific Christopher Wallis San Joaquin Valley Agricultural Sciences Center Follow this and additional works at: https://scholarlycommons.pacific.edu/cop-facarticles Part of the Chemistry Commons Recommended Citation Franz, A. H., Serebnitskaya, I., Gudial, G., & Wallis, C. (2014). Structure assignment and H/D-exchange behavior of several glycosylated polyphenols. ARKIVOC, 2014(5), 1–29. DOI: 10.3998/ark.5550190.p008.583 https://scholarlycommons.pacific.edu/cop-facarticles/143 This Article is brought to you for free and open access by the All Faculty Scholarship at Scholarly Commons. It has been accepted for inclusion in College of the Pacific aF culty Articles by an authorized administrator of Scholarly Commons. For more information, please contact [email protected]. General Papers ARKIVOC 2014 (v) 94-122 Structure assignment and H/D-exchange behavior of several glycosylated polyphenols Andreas H. Franz,a* Ilona Serebnitskaya,a Gurbir Gudial,a and Christopher Wallisb a Department of Chemistry, University of the Pacific, 3601 Pacific Avenue, Stockton, CA 95211, USA b Crop Diseases, Pests, and Genetics Research Unit, USDA-ARS San Joaquin Valley Agricultural Sciences Center, 9611 S. Riverbend Ave, Parlier, CA 93648, USA E-mail: [email protected] DOI: http://dx.doi.org/10.3998/ark.5550190.p008.583 Abstract The NMR-structures of six polyphenols, resveratrol (1), (-)-epicatechin (2), pelargonidin chloride (3), cyanidin chloride (4), cyanin chloride (5), and keracyanin chloride (6), were fully assigned.