Sarah Kreidler, PT, DPT, CSCS
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Oracle FLEXCUBE Core Banking Licensing Guide Release 11.5.0.0.0
Oracle FLEXCUBE Core Banking Licensing Guide Release 11.5.0.0.0 Part No. E52876-01 July 2014 Licensing Guide July 2014 Oracle Financial Services Software Limited Oracle Park Off Western Express Highway Goregaon (East) Mumbai, Maharashtra 400 063 India Worldwide Inquiries: Phone: +91 22 6718 3000 Fax:+91 22 6718 3001 www.oracle.com/financialservices/ Copyright © 2008, 2014, Oracle and/or its affiliates. All rights reserved. Oracle and Java are registered trademarks of Oracle and/or its affiliates. Other names may be trademarks of their respective owners. U.S. GOVERNMENT END USERS: Oracle programs, including any operating system, integrated software, any programs installed on the hardware, and/or documentation, delivered to U.S. Government end users are “commercial computer software” pursuant to the applicable Federal Acquisition Regulation and agency-specific supplemental regulations. As such, use, duplication, disclosure, modification, and adaptation of the programs, including any operating system, integrated software, any programs installed on the hardware, and/or documentation, shall be subject to license terms and license restrictions applicable to the programs. No other rights are granted to the U.S. Government. This software or hardware is developed for general use in a variety of information management applications. It is not developed or intended for use in any inherently dangerous applications, including applications that may create a risk of personal injury. If you use this software or hardware in dangerous applications, then you shall be responsible to take all appropriate failsafe, backup, redundancy, and other measures to ensure its safe use. Oracle Corporation and its affiliates disclaim any liability for any damages caused by use of this software or hardware in dangerous applications. -

Java Advanced Imaging
Java Advanced Imaging MIMUC Medientechnik SS 2003 Übersicht Wozu Bildbearbeitung mit Frameworks? Warum JAI? Verzahnung mit Java2D (AWT) JAI Packages und Typen Unterstützte Codecs und Bildformate BufferedImage, Raster, ColorModel (AWT) Rendergraphen Aufbau der Bildklassen Anzeigen eines gespeicherten Bildes Parameterblock Operationen Look-Up-Table Speichern eines Bildes Quellangabe MIMUC Medientechnik SS 2003 Helge Groß 2 - 23 Wozu Bildbearbeitung mit Frameworks? Rechner/Monitore können Bilder heute schnell verarbeiten/anzeigen Dadurch ist Analyse- und Bearbeitungsmöglichkeit am PC geschaffen Frameworks stellen bereit Bildmanipulationsmöglichkeiten Mathematische Operationen Bildanzeigemöglichkeit Bild-Codecs Einsatzgebiete Bildanalyse- und Bildanzeigesysteme Objektpositionierung, Objekttracking, Qualitätsprüfung Bildbearbeitungssysteme Medien, Werbeindustrie, Grafik-Design, Zeichenprogramme Applikationen Programme, Computerspiele World Wide Web Webseiten MIMUC Medientechnik SS 2003 Helge Groß 3 - 23 Warum JAI? Pro Plattformunabhängig Geringer Speicherplatz Schnelle Internetübertragung Applets im WWW Bildverarbeitende Web-Applikationen Als Frontend einer anderen Programmiersprache Diese übernimmt komplexere Operationen Client-Server-Architektur Komplexe Bildmanipulationen durchaus praktikabel Großer Leistungsumfang zur Bildverarbeitung Versucht alle Bedürfnisse zu erfüllen Extrem Erweiterbar Contra Langsam nicht zuletzt wegen Swing MIMUC Medientechnik SS 2003 Helge Groß 4 - 23 Verzahnung mit Java2D (AWT) JAI -
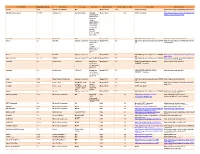
Third Party Version
Third Party Name Third Party Version Manufacturer License Type Comments Merge Product Merge Product Versions License details Software source autofac 3.5.2 Autofac Contributors MIT Merge Cardio 10.2 SOUP repository https://www.nuget.org/packages/Autofac/3.5 .2 Gibraltar Loupe Agent 2.5.2.815 eSymmetrix Gibraltor EULA Gibraltar Merge Cardio 10.2 SOUP repository https://my.gibraltarsoftware.com/Support/Gi Loupe Agent braltar_2_5_2_815_Download will be used within the Cardio Application to view events and metrics so you can resolve support issues quickly and easily. Modernizr 2.8.3 Modernizr MIT Merge Cadio 6.0 http://modernizr.com/license/ http://modernizr.com/download/ drools 2.1 Red Hat Apache License 2.0 it is a very old Merge PACS 7.0 http://www.apache.org/licenses/LICENSE- http://mvnrepository.com/artifact/drools/dro version of 2.0 ols-spring/2.1 drools. Current version is 6.2 and license type is changed too drools 6.3 Red Hat Apache License 2.0 Merge PACS 7.1 http://www.apache.org/licenses/LICENSE- https://github.com/droolsjbpm/drools/releases/ta 2.0 g/6.3.0.Final HornetQ 2.2.13 v2.2..13 JBOSS Apache License 2.0 part of JBOSS Merge PACS 7.0 http://www.apache.org/licenses/LICENSE- http://mvnrepository.com/artifact/org.hornet 2.0 q/hornetq-core/2.2.13.Final jcalendar 1.0 toedter.com LGPL v2.1 MergePacs Merge PACS 7.0 GNU LESSER GENERAL PUBLIC http://toedter.com/jcalendar/ server uses LICENSE Version 2. v1, and viewer uses v1.3. -

Programming in Java Advanced Imaging
Programming in Java™ Advanced Imaging Release 1.0.1 November 1999 JavaSoft A Sun Microsystems, Inc. Business 901 San Antonio Road Palo Alto, CA 94303 USA 415 960-1300 fax 415 969-9131 1999 Sun Microsystems, Inc. 901 San Antonio Road, Palo Alto, California 94303 U.S.A. All rights reserved. RESTRICTED RIGHTS LEGEND: Use, duplication, or disclosure by the United States Government is subject to the restrictions set forth in DFARS 252.227-7013 (c)(1)(ii) and FAR 52.227-19. The release described in this document may be protected by one or more U.S. patents, for- eign patents, or pending applications. Sun Microsystems, Inc. (SUN) hereby grants to you a fully paid, nonexclusive, nontrans- ferable, perpetual, worldwide limited license (without the right to sublicense) under SUN’s intellectual property rights that are essential to practice this specification. This license allows and is limited to the creation and distribution of clean-room implementa- tions of this specification that (i) are complete implementations of this specification, (ii) pass all test suites relating to this specification that are available from SUN, (iii) do not derive from SUN source code or binary materials, and (iv) do not include any SUN binary materials without an appropriate and separate license from SUN. Java, JavaScript, Java 3D, and Java Advanced Imaging are trademarks of Sun Microsys- tems, Inc. Sun, Sun Microsystems, the Sun logo, Java and HotJava are trademarks or reg- istered trademarks of Sun Microsystems, Inc. UNIX® is a registered trademark in the United States and other countries, exclusively licensed through X/Open Company, Ltd. -

CES Free Or Open Source Licenses Licenses Library Version 15.2 2020R1
CES Free or Open Source Licenses Licenses Library Version 15.2 2020R1 Revision 2.00 December 2020 Verint.com Twitter.com/verint Facebook.com/verint Blog.verint.com Table of Contents Free or Open Source Licenses ....................................................................................... 1 7-Zip - GNU LGPL + unRAR restrictions .................................................................... 1 ActivePython ............................................................................................................... 2 ANTLR .......................................................................................................................... 6 Apache License............................................................................................................ 6 ares Library................................................................................................................. 11 Attribution-NonCommercial-ShareAlike 3.0 Unported ............................................. 12 Batik SVG Toolkit ....................................................................................................... 17 Bouncy Castle ............................................................................................................ 19 Boost ........................................................................................................................... 20 BSD (4-Clause) License ............................................................................................ 20 COMMON DEVELOPMENT AND DISTRIBUTION -

OLS Evaluated Configuration for Oracle Database 10G Release 2 (10.2.0)
OLS Evaluated Configuration for Oracle Database 10g Release 2 (10.2.0) November 2007 Security Evaluations Oracle Corporation 500 Oracle Parkway Redwood Shores, CA 94065 OLS Evaluated Configuration for Oracle Database 10g i Release 2 (10.2.0) November 2007 Issue 0.6 OLS Evaluated Configuration for Oracle Database 10g Release 2 (10.2.0) November 2007 Author: Saad Syed, modifications made by Courtney Cavness Contributors: Peter Goatly, Shaun Lee This document is based on the equivalent document for OLS for Oracle10g Release 1, Issue 0.5 [OLSECD_10] used in the last Common Cri- teria Evaluation of Oracle10g with OLS. The contributions of the many authors of the precursors to this document are acknowledged. Change bars indicate changes made relative to [OLSECD_10]. Copyright © 1999, 2007, Oracle Corporation. All rights reserved.This documentation contains proprietary information of Oracle Corporation; it is protected by copyright law. Reverse engineering of the software is prohibited. If this documentation is delivered to a U.S. Government Agen- cy of the Department of Defense, then it is delivered with Restricted Rights and the following legend is applicable: RESTRICTED RIGHTS LEGEND Use, duplication or disclosure by the Government is subject to restrictions as set forth in subparagraph (c)(1)(ii) of DFARS 252.227-7013, Rights in Technical Data and Computer Software (October 1988). Oracle Corporation, 500 Oracle Parkway, Redwood City, CA 94065. The information in this document is subject to change without notice. If you find any problems in the documentation, please report them to us in writing. Oracle Corporation does not warrant that this document is error free. -
Oracle Database Licensing Information, 10G Release 2 (10.2) B14199-01
Oracle® Database Licensing Information 10g Release 2 (10.2) B14199-01 June 2005 Oracle Database Licensing Information, 10g Release 2 (10.2) B14199-01 Copyright © 2004, 2005, Oracle. All rights reserved. Contributors: Francisco Abedrabbo, Manmeet Ahluwalia, Sandra Cheevers, Anil Khilani, Mughees Minhas, Trish McGonigle, Dennis MacNeil, Anu Natarajan, Paul Needham, Jill Robinson, Mark Townsend, Jacqueline Woods The Programs (which include both the software and documentation) contain proprietary information; they are provided under a license agreement containing restrictions on use and disclosure and are also protected by copyright, patent, and other intellectual and industrial property laws. Reverse engineering, disassembly, or decompilation of the Programs, except to the extent required to obtain interoperability with other independently created software or as specified by law, is prohibited. The information contained in this document is subject to change without notice. If you find any problems in the documentation, please report them to us in writing. This document is not warranted to be error-free. Except as may be expressly permitted in your license agreement for these Programs, no part of these Programs may be reproduced or transmitted in any form or by any means, electronic or mechanical, for any purpose. If the Programs are delivered to the United States Government or anyone licensing or using the Programs on behalf of the United States Government, the following notice is applicable: U.S. GOVERNMENT RIGHTS Programs, software, databases, and related documentation and technical data delivered to U.S. Government customers are "commercial computer software" or "commercial technical data" pursuant to the applicable Federal Acquisition Regulation and agency-specific supplemental regulations. -

Evaluated Configuration for Oracle Database 10G Release 1 (10.1.0)
Evaluated Configuration for Oracle Database 10g Release 1 (10.1.0) November 2005 Security Evaluations Oracle Corporation 500 Oracle Parkway Redwood Shores, CA 94065 Evaluated Configuration for Oracle Database 10g i Release 1 (10.1.0) November 2005 Issue 0.5 Evaluated Configuration for Oracle Database 10g Release 1 (10.1.0) November 2005 Author: Saad Syed Contributors: Peter Goatly, Shaun Lee This document is based on the equivalent document for Oracle9i Release 2, Issue 1.0 [ECD_10] used in the last Common Criteria Evaluation of Oracle9i. The contributions of the many authors of the precursors to this document are acknowledged. Change bars indicate changes made relative to [ECD_10]. Copyright © 1999, 2005, Oracle Corporation. All rights reserved.This documentation contains proprietary information of Oracle Corporation; it is protected by copyright law. Reverse engineering of the software is prohibited. If this documentation is delivered to a U.S. Government Agen- cy of the Department of Defense, then it is delivered with Restricted Rights and the following legend is applicable: RESTRICTED RIGHTS LEGEND Use, duplication or disclosure by the Government is subject to restrictions as set forth in subparagraph (c)(1)(ii) of DFARS 252.227-7013, Rights in Technical Data and Computer Software (October 1988). Oracle Corporation, 500 Oracle Parkway, Redwood City, CA 94065. The information in this document is subject to change without notice. If you find any problems in the documentation, please report them to us in writing. Oracle Corporation does not warrant that this document is error free. Oracle is a registered trademark and Oracle Database 10g, Oracle9i, PL/SQL, Oracle Enterprise Manager, Oracle Call Interface, SQL*Plus, SQL*Loader, Oracle Net and Oracle Label Security are trademarks or registered trademarks of Oracle Corporation. -

An Integrated Solution to Display Large Images at Full Resolution
4th Planetary Data Workshop 2019 (LPI Contrib. No. 2151) 7064.pdf AN INTEGRATED SOLUTION TO DISPLAY LARGE IMAGES AT FULL RESOLUTION. J. E. Turner1, F. S. Turner1, C. M. O’Shea1, and G. W. Patterson1. Johns Hopkins University Applied Physics Laboratory, 11100 Johns Hopkins Rd, Laurel, MD 20723 ([email protected]) Introduction: As technology advances, imaging tiling method loads only the visible portion of the image instruments are becoming more capable and into memory, and is the key to loading overbearingly increasingly higher resolution. Consequently, these large images. instruments are recording more data than ever before, As the tiles required by the display are loaded from and multi-gigabyte files are now common place. Along JAI, they are potentially stored into a configurable with images becoming larger and higher resolution, the memory cache. This eliminates the need to recreate processes applied to imaging data have also become some subset of tiles when the display changes, thus more complex. increasing rendering speed. Alternatively, disk-based With this advancement in imaging technology cache implementations also exist. The features of JAI comes challenges with loading and displaying these enable complex operational chains to be constructed extremely large data sets. Some images are so large that with little or no computational effort. The API supports loading them into RAM effectively kills a computer’s multi-threaded computation so the algorithms can take interactivity. We are working to develop a software advantage of the parallel capabilities of multi-core framework that can load these large imaging datasets processors common in most computers today. There is efficiently, apply complex processing algorithms to an inherent 2-gigapixel limit in the more common push- them, and easily visualize the results. -

Analyzing Huge Pathology Images with Open Source Software
Analyzing huge pathology images with open source software Christophe Deroulers∗1, David Ameisen2 , Mathilde Badoual1 , Chlo´eGerin3;4 , Alexandre Granier5 and Marc Lartaud5;6 1Univ Paris Diderot, Laboratoire IMNC, UMR 8165 CNRS, Univ Paris-Sud, F-91405 Orsay, France 2Univ Paris Diderot, Laboratoire de pathologie, H^opitalSaint-Louis APHP, INSERM UMR-S 728, F-75010 Paris, France 3on leave from CNRS, UMR 8165, Laboratoire IMNC, Univ Paris-Sud, Univ Paris Diderot, F-91405 Orsay, France 4now at CNRS, UMR 8148, Laboratoire IDES, Univ Paris-Sud, F-91405 Orsay, France and CNRS, UMR 8608, IPN, Univ Paris-Sud, F-91405 Orsay, France 5MRI-Montpellier RIO Imaging, CRBM, F-34293 Montpellier, France 6CIRAD, F-34398 Montpellier CEDEX 5, France Email: Christophe Deroulers∗- [email protected]; David Ameisen - [email protected]; Mathilde Badoual - [email protected]; Chlo´eGerin - [email protected]; Alexandre Granier - [email protected]; Marc Lartaud - [email protected]; ∗Corresponding author Abstract Background: Digital pathology images are increasingly used both for diagnosis and research, because slide scanners are nowadays broadly available and because the quantitative study of these images yields new insights in systems biology. However, such virtual slides build up a technical challenge since the images occupy often several gigabytes and cannot be fully opened in a computer's memory. Moreover, there is no standard format. Therefore, most common open source tools such as ImageJ fail at treating them, and the others require expensive hardware while still being prohibitively slow. Results: We have developed several cross-platform open source software tools to overcome these limitations. -

Building VICAR Open Source Version Version 2.0
Building VICAR Open Source version Version 2.0 2016-05-22 Prepared by: _________________________ Walter Bunch _________________________ Robert Deen, VICAR Cognizant Engineer Jet Propulsion Laboratory California Institute of Technology Pasadena, California Copyright 2016 California Institute of Technology. Government sponsorship acknowledged. Building VICAR 1. Introduction............................................................................................................................................... 3 1.1. Supported Platforms ................................................................................................................................... 3 2. Obtaining/Installing VICAR .................................................................................................................. 4 2.1. Obtaining VICAR ........................................................................................................................................... 4 2.2. Directory Layout .......................................................................................................................................... 4 2.2.1. Installing a Pre-Built VICAR...................................................................................................................................... 5 3. External Libraries .................................................................................................................................... 6 3.1. commons-vfs ................................................................................................................................................ -

Imvision OSS Open Source Software
imVision® Open Source Software 860641279 Revision C June 2019 commscope.com NOTE: This document revision applies to imVision® System Manager version 8.2. To obtain open source software license information for subsequent versions of imVision® System Manager, refer to the “Legal Notices” section of imVision® System Manager’s help system. imVision® System Manager Open Source Software (OSS) Catalog Table of Contents Purpose and Scope ....................................................................................................................................... 2 Open Source Software Catalog Table ........................................................................................................... 2 Licenses ........................................................................................................................................................ 6 Apache License Version 2.0 ...................................................................................................................... 6 BSD ............................................................................................................................................................ 8 COMMON DEVELOPMENT AND DISTRIBUTION LICENSE (CDDL v1.0) ............................................. 9 Eclipse Public License - v1.0 ................................................................................................................... 15 GNU Lesser General Public License v2.1 ..............................................................................................