Fasting-Induced FGF21 Signaling Activates Hepatic Autophagy and Lipid Degradation Via JMJD3 Histone Demethylase
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Download File
HHS Public Access Author manuscript Author Manuscript Author ManuscriptAm J Med Author Manuscript Genet B Neuropsychiatr Author Manuscript Genet. Author manuscript; available in PMC 2017 January 01. Published in final edited form as: Am J Med Genet B Neuropsychiatr Genet. 2016 January ; 171(1): 44–64. doi:10.1002/ajmg.b.32384. Genetic Influences on the Neural and Physiological Bases of Acute Threat: A Research Domain Criteria (RDoC) Perspective Jennifer A. Sumner1,2, Abigail Powers3, Tanja Jovanovic3, and Karestan C. Koenen2,4,5 1Center for Behavioral Cardiovascular Health, Columbia University Medical Center, New York, NY 2Department of Epidemiology, Harvard T.H. Chan School of Public Health, Boston, MA 3Department of Psychiatry and Behavioral Sciences, Emory University School of Medicine, Atlanta, GA 4Psychiatric and Neurodevelopmental Genetics Unit and Department of Psychiatry, Massachusetts General Hospital, Boston, MA 5The Stanley Center for Psychiatric Research at the Broad Institute of MIT and Harvard, Cambridge, MA Abstract The NIMH Research Domain Criteria (RDoC) initiative aims to describe key dimensional constructs underlying mental function across multiple units of analysis—from genes to observable behaviors—in order to better understand psychopathology. The acute threat (“fear”) construct of the RDoC Negative Valence System has been studied extensively from a translational perspective, and is highly pertinent to numerous psychiatric conditions, including anxiety and trauma-related disorders. We examined genetic contributions to the construct of acute threat at two units of analysis within the RDoC framework: 1) neural circuits and 2) physiology. Specifically, we focused on genetic influences on activation patterns of frontolimbic neural circuitry and on startle, skin conductance, and heart rate responses. -

Review Article Redundant Kinase Activation and Resistance of EGFR-Tyrosine Kinase Inhibitors
Am J Cancer Res 2014;4(6):608-628 www.ajcr.us /ISSN:2156-6976/ajcr0001942 Review Article Redundant kinase activation and resistance of EGFR-tyrosine kinase inhibitors Min Luo, Li-Wu Fu Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, 510060, China Received August 17, 2014; Accepted October 12, 2014; Epub November 19, 2014; Published November 30, 2014 Abstract: Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) have shown dramatic effects against that tumors harboring EGFR activating mutations in the EGFR intracytoplasmic tyrosine kinase domain and resulted in cell apoptosis. Unfortunately, a number of patients ultimately developed resistance by multiple mechanisms. Thus, elucidation of the mechanism of resistance to EGFR-TKIs can provide strategies for blocking or reversing the situation. Recent studies suggested that redundant kinase activation plays pivotal roles in escaping from the effects of EGFR-TKIs. Herein, we aimed to characterize several molecular events involved in the resistance to EGFR-TKIs mediated by redundant kinase activation. Keywords: EGFR, redundant kinase activation, resistance to EGFR-TKIs Introduction ligand-binding extracellular domain, thereby preventing ligand binding, and consequently re- Epidermal growth factor receptor (EGFR), a ceptor dimerization, and receptor signaling. member of a family which consists of at least 4 Among these, gefitinib and erlotinib were the receptor tyrosine kinases, including EGFR (Er- first EGFR-TKIs to be approved by Food and bB1), HER2 (ErbB2), HER3 (ErbB3), and HER4 Drug Administration (FDA) for treatment of (ErbB4) (Figure 1). To date, seven ligands for NSCLC (Table 1). -

Cold-Induced Epigenetic Programming of the Sperm Enhances Brown Adipose Tissue Activity in the Offspring
ARTICLES https://doi.org/10.1038/s41591-018-0102-y Cold-induced epigenetic programming of the sperm enhances brown adipose tissue activity in the offspring Wenfei Sun1,9, Hua Dong1,9, Anton S. Becker 1,2,3, Dianne H. Dapito1, Salvatore Modica1, Gerald Grandl1, Lennart Opitz1,4, Vissarion Efthymiou1, Leon G. Straub1, Gitalee Sarker1, Miroslav Balaz1, Lucia Balazova1, Aliki Perdikari1, Elke Kiehlmann1, Sara Bacanovic3, Caroline Zellweger3, Daria Peleg-Raibstein1, Pawel Pelczar5, Wolf Reik6,7, Irene A. Burger3, Ferdinand von Meyenn 6,8 and Christian Wolfrum1* Recent research has focused on environmental effects that control tissue functionality and systemic metabolism. However, whether such stimuli affect human thermogenesis and body mass index (BMI) has not been explored. Here we show retrospec- tively that the presence of brown adipose tissue (BAT) and the season of conception are linked to BMI in humans. In mice, we demonstrate that cold exposure (CE) of males, but not females, before mating results in improved systemic metabolism and protection from diet-induced obesity of the male offspring. Integrated analyses of the DNA methylome and RNA sequencing of the sperm from male mice revealed several clusters of co-regulated differentially methylated regions (DMRs) and differentially expressed genes (DEGs), suggesting that the improved metabolic health of the offspring was due to enhanced BAT formation and increased neurogenesis. The conclusions are supported by cell-autonomous studies in the offspring that demonstrate an enhanced capacity to form mature active brown adipocytes, improved neuronal density and more norepinephrine release in BAT in response to cold stimulation. Taken together, our results indicate that in humans and in mice, seasonal or experimental CE induces an epigenetic programming of the sperm such that the offspring harbor hyperactive BAT and an improved adaptation to overnutrition and hypothermia. -

EGFR in Cancer: Signaling Mechanisms, Drugs, and Acquired Resistance
cancers Review EGFR in Cancer: Signaling Mechanisms, Drugs, and Acquired Resistance Mary Luz Uribe , Ilaria Marrocco and Yosef Yarden * Department of Biological Regulation, Weizmann Institute of Science, Rehovot 7610001, Israel; [email protected] (M.L.U.); [email protected] (I.M.) * Correspondence: [email protected] Simple Summary: Growth factors are hormone-like molecules able to promote division and mi- gration of normal cells, but cancer captured the underlying mechanisms to unleash tumor growth and metastasis. Here we review the epidermal growth factor (EGF), which controls epithelial cells, the precursors of all carcinomas, and the cognate cell surface receptor, called EGFR. In addition to over-production of EGF and its family members in tumors, EGFR is similarly over-produced, and mutant hyper-active forms of EGFR are uniquely found in some brain, lung, and other cancers. After describing the biochemical mechanisms underlying the cancer-promoting actions of EGFR, we review some of the latest research discoveries and list all anti-cancer drugs specifically designed to block the EGFR’s biochemical pathway. We conclude by explaining why some patients with lung or colorectal cancer do not respond to the anti-EGFR therapies and why still other patients, who initially respond, become tolerant to the drugs. Abstract: The epidermal growth factor receptor (EGFR) has served as the founding member of the large family of growth factor receptors harboring intrinsic tyrosine kinase function. High abundance of EGFR and large internal deletions are frequently observed in brain tumors, whereas Citation: Uribe, M.L.; Marrocco, I.; point mutations and small insertions within the kinase domain are common in lung cancer. -

Fibroblast Growth Factor Receptor Is a Mechanistic Link Between Visceral Adiposity and Cancer
OPEN Oncogene (2017), 1–12 www.nature.com/onc ORIGINAL ARTICLE Fibroblast growth factor receptor is a mechanistic link between visceral adiposity and cancer D Chakraborty1, V Benham1, B Bullard1, T Kearney2, HC Hsia3, D Gibbon4, EY Demireva5, SY Lunt6 and JJ Bernard1 Epidemiological evidence implicates excess adipose tissue in increasing cancer risk. Despite a steeply rising global prevalence of obesity, how adiposity contributes to transformation (stage a non-tumorigenic cell undergoes to become malignant) is unknown. To determine the factors in adipose tissue that stimulate transformation, we used a novel ex vivo system of visceral adipose tissue (VAT)-condition medium-stimulated epithelial cell growth in soft agar. To extend this system in vivo, we used a murine lipectomy model of ultraviolet light B-induced, VAT-promoted skin tumor formation. We found that VAT from mice and obese human donors stimulated growth in soft agar of non-tumorigenic epithelial cells. The difference in VAT activity was associated with fibroblast growth factor-2 (FGF2) levels. Moreover, human and mouse VAT failed to stimulate growth in soft of agar in cells deficient in FGFR-1 (FGF2 receptor). We also demonstrated that circulating levels of FGF2 were associated with non-melanoma tumor formation in vivo. These data implicate FGF2 as a major factor VAT releases to transform epithelial cells—a novel, potential pathway of VAT-enhanced tumorigenesis. Strategies designed to deplete VAT stores of FGF2 or inhibit FGFR-1 in abdominally obese individuals may be important cancer prevention strategies as well as adjuvant therapies for improving outcomes. Oncogene advance online publication, 7 August 2017; doi:10.1038/onc.2017.278 INTRODUCTION adiposity is independently and positively associated with cancer 14 The carcinogenic impact of having excess adipose tissue is risk in older men. -

C-Src Activation Mediates Erlotinib Resistance in Head and Neck Cancer by Stimulating C-Met
Published OnlineFirst December 4, 2012; DOI: 10.1158/1078-0432.CCR-12-1555 Clinical Cancer Cancer Therapy: Preclinical Research c-Src Activation Mediates Erlotinib Resistance in Head and Neck Cancer by Stimulating c-Met Laura P. Stabile1, Guoqing He3, Vivian Wai Yan Lui2, Cassandra Henry1, Christopher T. Gubish1, Sonali Joyce2, Kelly M. Quesnelle2, Jill M. Siegfried1, and Jennifer R. Grandis1,2 Abstract Purpose: Strategies to inhibit the EGF receptor (EGFR) using the tyrosine kinase inhibitor erlotinib have been associated with limited clinical efficacy in head and neck squamous cell carcinoma (HNSCC). Co-activation of alternative kinases may contribute to erlotinib resistance. Experimental Design: We generated HNSCC cells expressing dominant-active c-Src (DA-Src) to determine the contribution of c-Src activation to erlotinib response. Results: Expression of DA-Src conferred resistance to erlotinib in vitro and in vivo compared with vector- transfected control cells. Phospho-Met was strongly upregulated by DA-Src, and DA-Src cells did not produce hepatocyte growth factor (HGF). Knockdown of c-Met enhanced sensitivity to erlotinib in DA-Src cells in vitro, as did combining a c-Met or c-Src inhibitor with erlotinib. Inhibiting EGFR resulted in minimal reduction of phospho-Met in DA-Src cells, whereas complete phospho-Met inhibition was achieved by inhibiting c-Src. A c-Met inhibitor significantly sensitized DA-Src tumors to erlotinib in vivo, resulting in reduced Ki67 labeling and increased apoptosis. In parental cells, knockdown of endogenous c-Src enhanced sensitivity to erlotinib, whereas treatment with HGF to directly induce phospho-Met resulted in erlotinib resistance. -

Novel Epigenetic and Genetic Biomarker Candidates in Post-Traumatic Stress Disorder
Dissertation zur Erlangung des Doktorgrades der Fakultät für Biologie der Ludwig-Maximilians-Universität München Novel Epigenetic and Genetic Biomarker Candidates in Post-Traumatic Stress Disorder Vorgelegt von Julius-Christopher Pape aus München 2020 Novel epigenetic and genetic biomarker candidates in PTSD Tag der Einreichung: 17.09.2020 Tag der mündlichen Prüfung: 16.04.2021 Erster Gutachter: PD Dr. Mathias V. Schmidt Zweiter Gutachter: Prof. Dr. Laura Busse Dritter Gutachter: Prof. Dr. Anja Horn-Bochtler Vierter Gutachter: Prof. Dr. Peter Geigenberger Fünfter Gutachter: Prof. Dr. Wolfgang Enard Sechster Gutachter: Prof. Dr. Heinrich Leonhardt 2 Novel epigenetic and genetic biomarker candidates in PTSD Content Page List of Abbreviations 4 List of Publications 6 Summary 7 Zusammenfassung 9 Introduction 12 Aims of the thesis 35 Paper I, Pape et al., Clinical Epigenetics, 2018, 10(1):136 36 Paper II, Dunlop et al., Biological Psychiatry, 2017,82(12):866-874 48 Paper III, Volk et al., Cell Reports, 2016, 17(7):1882.1891 58 Discussion 70 References 87 Supplemental materials • Paper I 100 • Paper II 102 • Paper III 115 Addendum, Pape & Binder, Psychiatric Annals, 2016, 46(9):510-518 126 Acknowledgements 136 Curriculum Vitae 137 Eidesstattliche Erklärung 139 Declaration of contribution/as co-author 140 3 Novel epigenetic and genetic biomarker candidates in PTSD Abbreviations 5-HTTLPR Serotonin-transporter-linked polymorphic region ACTH Adrenocorticotropic hormone ADCYAP1R1 Pituitary adenylate cyclase-activating polypeptide type I receptor -

Ectodomain Shedding of the EGF-Receptor Ligand Epigen Is Mediated by ADAM17
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector FEBS Letters 581 (2007) 41–44 Ectodomain shedding of the EGF-receptor ligand epigen is mediated by ADAM17 Umut Sahina, Carl P. Blobela,b,* a Arthritis and Tissue Degeneration Program, Caspary Research Building, Room 426, Hospital for Special Surgery, 535 East 70th Street, New York, NY 10021, USA b Departments of Medicine and of Physiology and Biophysics, Weill Medical College of Cornell University, New York, NY 10021, USA Received 26 October 2006; accepted 26 November 2006 Available online 6 December 2006 Edited by Richard Marais concentrations of EGF, TGFa or epigen, epigen was found Abstract All ligands of the epidermal growth factor receptor (EGFR), which has important roles in development and disease, to be the most potent mitogen [12]. Epigen effectively activates are made as transmembrane precursors. Proteolytic processing signaling via an EGFR-homodimer or through a heterodimer by ADAMs (a disintegrin and metalloprotease) regulates the bio- between EGFR and ErbB2 [12]. Epigen is widely expressed in availability of several EGFR-ligands, yet little is known about a variety of mouse tissues, especially in multiple developing the enzyme responsible for processing the recently identified structures in the embryo, such as developing tongue papillae, EGFR ligand, epigen. Here we show that ectodomain shedding the dorsal root ganglion and the inner and outer root sheath of epigen requires ADAM17, which can be stimulated by phorbol of hair follicles, all of which are regions of active proliferation esters, phosphatase inhibitors and calcium influx. -
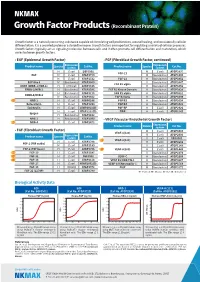
Growth Factor Products (Recombinant Protein)
Growth Factor Products (Recombinant Protein) Growth factor is a naturally occurring substance capable of stimulating cell proliferation, wound healing, and occasionally cellular differentiation. It is a secreted protein or a steroid hormone. Growth factors are important for regulating a variety of cellular processes. Growth factors typically act as signaling molecules between cells and it often promote cell differentiation and maturation, which varies between growth factors. • EGF (Epidermal Growth Factor) • FGF (Fibroblast Growth Factor, continued) Expression Expression Product name Species system Cat No. Product name Species system Cat No. H E.coli EGF0501 M E.coli ATGP3897 FGF-21 EGF M E.coli ATGP3729 M Baculovirus ATGP3300 M E.coli ATGP1532 FGF-23 H Baculovirus ATGP3852 EGF-like 6 M Baculovirus ATGP3860 H Baculovirus ATGP3647 FGF R1 alpha EGFR (ERBB-1/HER-1) H CHO Cell ATGP3335 H Baculovirus ATGP3856 ERBB-2/HER-2 H Baculovirus ATGP3586 FGF R1 Kinase Domain H Baculovirus ATGP3828 H Baculovirus ATGP3125 FGF R2 alpha H Baculovirus ATGP3621 ERBB-3/HER-3 M Baculovirus ATGP3302 FGF R2 beta H Baculovirus ATGP3899 GRB-2 H E.coli ATGP0286 FGF R3 H Baculovirus ATGP3654 Betacellulin H E.coli ATGP1250 FGF R4 H Baculovirus ATGP3423 Epiregulin H E.coli ATGP2923(D) FGF-BP H E.coli ATGP2926 H E.coli ATGP2332(D) FIBP H E.coli ATGP2145 Epigen H Baculovirus ATGP3682 NRG-1 H Baculovirus ATGP3990 • VEGF (Vascular Endothelial Growth Factor) NRG-4 H E.coli ATGP2472 Expression Product name Species system Cat No. • FGF (Fibroblast Growth Factor) M E.coli ATGP0389 VEGF-A(120) R E.coli ATGP2900 Product name Species Expression Cat No. -

Product Sheet CP10075
EGF Antibody Applications: WB, IP, ELISA, ICC, IHC Detected MW: 6.2 kDa Cat. No. CP10075 Species & Reactivity: Human, Mouse Isotype: Mouse IgG1 BACKGROUND APPLICATIONS EGF is the founding member of the EGF-family of Application: *Dilution: protein. Members of this protein family have highly WB 1:1000 similar structural and functional characteristics. IP 1:50 Human EGF is a 6045-Da protein with 53 amino IHC 1:200 acid residues and three intramolecular disulfide ICC 1:200 bonds. Besides EGF itself other family members include: HB-EGF, TGF-α, Amphiregulin, Epiregulin, ELISA 1:10,000 *Optimal dilutions must be determined by end user. Epigen, Betacellulin, NRG-1, -2, -3, and -4. EGF has a profound effect on the differentiation of specific cells in vivo and is a potent mitogenic factor for a variety of cultured cells of both QUALITY CONTROL DATA ectodermal and mesodermal origin. The EGF precursor is believed to exist as a membrane- bound molecule which is proteolytically cleaved to generate the 53-amino acid peptide hormone that stimulates cells to divide. EGF stimulates the growth of various epidermal and epithelial tissues in vivo and in vitro and of some fibroblasts in cell culture.1 EGF acts by binding with high affinity to epidermal growth factor receptor (EGFR) on the cell surface and stimulating the intrinsic protein-tyrosine kinase activity of the receptor. The tyrosine kinase activity, in turn, initiates a signal transduction cascade that results in a variety of biochemical changes within the cell, a rise in intracellular calcium levels, increased glycolysis and protein synthesis, and increases in the expression of certain genes including the gene for EGFR - that ultimately lead to DNA synthesis and cell proliferation.2 References: 1. -

ADAM10 and ADAM17 Regulate EGFR, C-Met and TNF RI Signalling In
www.nature.com/scientificreports OPEN ADAM10 and ADAM17 regulate EGFR, c‑Met and TNF RI signalling in liver regeneration and fbrosis Olga Zbodakova1*, Karel Chalupsky1,2, Lenka Sarnova1,3, Petr Kasparek1,2, Marketa Jirouskova1,3, Martin Gregor1,3 & Radislav Sedlacek 1,2* ADAM10 and ADAM17 are proteases that afect multiple signalling pathways by releasing molecules from the cell surface. As their substrate specifcities partially overlaps, we investigated their concurrent role in liver regeneration and fbrosis, using three liver‑specifc defcient mouse lines: ADAM10‑ and ADAM17‑defcient lines, and a line defcient for both proteases. In the model of partial hepatectomy, double defcient mice exhibited decreased AKT phosphorylation, decreased release of EGFR activating factors and lower shedding of HGF receptor c‑Met. Thus, simultaneous ablation of ADAM10 and ADAM17 resulted in inhibited EGFR signalling, while HGF/c‑Met signalling pathway was enhanced. In contrast, antagonistic efects of ADAM10 and ADAM17 were observed in the model of chronic CCl4 intoxication. While ADAM10‑defcient mice develop more severe fbrosis manifested by high ALT, AST, ALP and higher collagen deposition, combined defciency of ADAM10 and ADAM17 surprisingly results in comparable degree of liver damage as in control littermates. Therefore, ADAM17 defciency is not protective in fbrosis development per se, but can ameliorate the damaging efect of ADAM10 defciency on liver fbrosis development. Furthermore, we show that while ablation of ADAM17 resulted in decreased shedding of TNF RI, ADAM10 defciency leads to increased levels of soluble TNF RI in serum. In conclusion, hepatocyte‑derived ADAM10 and ADAM17 are important regulators of growth receptor signalling and TNF RI release, and pathological roles of these proteases are dependent on the cellular context. -
Advertising (PDF)
santa cruz biotechnology, inc. The Power to Question siRNA gene silencers and immunodetection reagents SEE OUR WEB SITE FEATURING: 11,915 monoclonal antibodies 19,623 polyclonal antibodies 13,269 siRNAs and support products HEADQUARTERS EUROPEAN SUPPORT ASIAN SUPPORT TEL: 831.457.3800 TEL: +49.6221.4503.0 JAPAN TOLL FREE: (010) 800.40402026 TOLL FREE: 1.800.457.3801 TOLL FREE: +00800.4573.8000 S. KOREA TOLL FREE: 00798.1.1.002.0297 FAX: 831.457.3801 FAX: +49.6221.4503.45 CHINA TOLL FREE: 10.800.711.0752 or 10.800.110.0694 EMAIL: [email protected] EMAIL: [email protected] EMAIL: [email protected] © 2006-2007 Santa Cruz Biotechnology, Inc., all rights reserved. "Santa Cruz Biotechnology" and the Santa Cruz Biotechnology, Inc. logo are registered trademarks of Santa Cruz Biotechnology, Inc. The Standard of Excellence -- Rely on eBioscience to Enhance Your Rate of Discovery Hey! Have you heard that eBioscience has launched PerCP-Cy5.5 conjugates? Really?! Finally, another choice. Data Comparison Specificities Histogram Overlay HUMAN MOUSE CD3 60 CD3 CD4 CD4 40 CD8 CD8 20 CD14 CD19 CD19 0 0 1 2 3 4 10 10 10 10 10 Counts CD20 CD45 eBioscience Leading Competitor CD25 CD45R/B220 60 60 CD33 CD90 40 40 CD45 20 20 CD127 CD127 γ 0 0 IFN- 0 1 2 3 4 0 1 2 3 4 γ 10 10 10 10 10 10 10 10 10 10 IFN- PerCP-Cy5.5 - FL3 Streptavidin Ly-6G/Gr1 The choice is clear - eBioscience For more details and to see other new products, visit www.ebioscience.com/PerCP-Cy55/JI.