Expression of Primary Cilia-Related Genes in Developing Mouse Gonads R.P
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Mea6 Controls VLDL Transport Through the Coordinated Regulation of COPII Assembly
Cell Research (2016) 26:787-804. npg © 2016 IBCB, SIBS, CAS All rights reserved 1001-0602/16 $ 32.00 ORIGINAL ARTICLE www.nature.com/cr Mea6 controls VLDL transport through the coordinated regulation of COPII assembly Yaqing Wang1, *, Liang Liu1, 2, *, Hongsheng Zhang1, 2, Junwan Fan1, 2, Feng Zhang1, 2, Mei Yu3, Lei Shi1, Lin Yang1, Sin Man Lam1, Huimin Wang4, Xiaowei Chen4, Yingchun Wang1, Fei Gao5, Guanghou Shui1, Zhiheng Xu1, 6 1State Key Laboratory of Molecular Developmental Biology, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China; 2University of Chinese Academy of Sciences, Beijing 100101, China; 3School of Life Science, Shandong University, Jinan 250100, China; 4Institute of Molecular Medicine, Peking University, Beijing 100871, China; 5State Key Laboratory of Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; 6Translation- al Medical Center for Stem Cell Therapy, Shanghai East Hospital, Tongji University School of Medicine, Shanghai 200120, China Lipid accumulation, which may be caused by the disturbance in very low density lipoprotein (VLDL) secretion in the liver, can lead to fatty liver disease. VLDL is synthesized in endoplasmic reticulum (ER) and transported to Golgi apparatus for secretion into plasma. However, the underlying molecular mechanism for VLDL transport is still poor- ly understood. Here we show that hepatocyte-specific deletion of meningioma-expressed antigen 6 (Mea6)/cutaneous T cell lymphoma-associated antigen 5C (cTAGE5C) leads to severe fatty liver and hypolipemia in mice. Quantitative lip- idomic and proteomic analyses indicate that Mea6/cTAGE5 deletion impairs the secretion of different types of lipids and proteins, including VLDL, from the liver. -

Mechanical Forces Induce an Asthma Gene Signature in Healthy Airway Epithelial Cells Ayşe Kılıç1,10, Asher Ameli1,2,10, Jin-Ah Park3,10, Alvin T
www.nature.com/scientificreports OPEN Mechanical forces induce an asthma gene signature in healthy airway epithelial cells Ayşe Kılıç1,10, Asher Ameli1,2,10, Jin-Ah Park3,10, Alvin T. Kho4, Kelan Tantisira1, Marc Santolini 1,5, Feixiong Cheng6,7,8, Jennifer A. Mitchel3, Maureen McGill3, Michael J. O’Sullivan3, Margherita De Marzio1,3, Amitabh Sharma1, Scott H. Randell9, Jefrey M. Drazen3, Jefrey J. Fredberg3 & Scott T. Weiss1,3* Bronchospasm compresses the bronchial epithelium, and this compressive stress has been implicated in asthma pathogenesis. However, the molecular mechanisms by which this compressive stress alters pathways relevant to disease are not well understood. Using air-liquid interface cultures of primary human bronchial epithelial cells derived from non-asthmatic donors and asthmatic donors, we applied a compressive stress and then used a network approach to map resulting changes in the molecular interactome. In cells from non-asthmatic donors, compression by itself was sufcient to induce infammatory, late repair, and fbrotic pathways. Remarkably, this molecular profle of non-asthmatic cells after compression recapitulated the profle of asthmatic cells before compression. Together, these results show that even in the absence of any infammatory stimulus, mechanical compression alone is sufcient to induce an asthma-like molecular signature. Bronchial epithelial cells (BECs) form a physical barrier that protects pulmonary airways from inhaled irritants and invading pathogens1,2. Moreover, environmental stimuli such as allergens, pollutants and viruses can induce constriction of the airways3 and thereby expose the bronchial epithelium to compressive mechanical stress. In BECs, this compressive stress induces structural, biophysical, as well as molecular changes4,5, that interact with nearby mesenchyme6 to cause epithelial layer unjamming1, shedding of soluble factors, production of matrix proteins, and activation matrix modifying enzymes, which then act to coordinate infammatory and remodeling processes4,7–10. -

Hras Intracellular Trafficking and Signal Transduction Jodi Ho-Jung Mckay Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 2007 HRas intracellular trafficking and signal transduction Jodi Ho-Jung McKay Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Biological Phenomena, Cell Phenomena, and Immunity Commons, Cancer Biology Commons, Cell Biology Commons, Genetics and Genomics Commons, and the Medical Cell Biology Commons Recommended Citation McKay, Jodi Ho-Jung, "HRas intracellular trafficking and signal transduction" (2007). Retrospective Theses and Dissertations. 13946. https://lib.dr.iastate.edu/rtd/13946 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. HRas intracellular trafficking and signal transduction by Jodi Ho-Jung McKay A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Genetics Program of Study Committee: Janice E. Buss, Co-major Professor Linda Ambrosio, Co-major Professor Diane Bassham Drena Dobbs Ted Huiatt Iowa State University Ames, Iowa 2007 Copyright © Jodi Ho-Jung McKay, 2007. All rights reserved. UMI Number: 3274881 Copyright 2007 by McKay, Jodi Ho-Jung All rights reserved. UMI Microform 3274881 Copyright 2008 by ProQuest Information and Learning Company. All rights reserved. This microform edition is protected against unauthorized copying under Title 17, United States Code. ProQuest Information and Learning Company 300 North Zeeb Road P.O. -

RNF11 at the Crossroads of Protein Ubiquitination
biomolecules Review RNF11 at the Crossroads of Protein Ubiquitination Anna Mattioni, Luisa Castagnoli and Elena Santonico * Department of Biology, University of Rome Tor Vergata, Via della ricerca scientifica, 00133 Rome, Italy; [email protected] (A.M.); [email protected] (L.C.) * Correspondence: [email protected] Received: 29 September 2020; Accepted: 8 November 2020; Published: 11 November 2020 Abstract: RNF11 (Ring Finger Protein 11) is a 154 amino-acid long protein that contains a RING-H2 domain, whose sequence has remained substantially unchanged throughout vertebrate evolution. RNF11 has drawn attention as a modulator of protein degradation by HECT E3 ligases. Indeed, the large number of substrates that are regulated by HECT ligases, such as ITCH, SMURF1/2, WWP1/2, and NEDD4, and their role in turning off the signaling by ubiquitin-mediated degradation, candidates RNF11 as the master regulator of a plethora of signaling pathways. Starting from the analysis of the primary sequence motifs and from the list of RNF11 protein partners, we summarize the evidence implicating RNF11 as an important player in modulating ubiquitin-regulated processes that are involved in transforming growth factor beta (TGF-β), nuclear factor-κB (NF-κB), and Epidermal Growth Factor (EGF) signaling pathways. This connection appears to be particularly significant, since RNF11 is overexpressed in several tumors, even though its role as tumor growth inhibitor or promoter is still controversial. The review highlights the different facets and peculiarities of this unconventional small RING-E3 ligase and its implication in tumorigenesis, invasion, neuroinflammation, and cancer metastasis. Keywords: Ring Finger Protein 11; HECT ligases; ubiquitination 1. -

S100A8 Transported by SEC23A Inhibits Metastatic Colonization Via
Sun et al. Cell Death and Disease (2020) 11:650 https://doi.org/10.1038/s41419-020-02835-w Cell Death & Disease ARTICLE Open Access S100A8 transported by SEC23A inhibits metastatic colonization via autocrine activation of autophagy Zhiwei Sun1,2, Bin Zeng1,2,DoudouLiu1,2, Qiting Zhao1,2,JianyuWang1,2 and H. Rosie Xing2,3 Abstract Metastasis is the main cause of failure of cancer treatment. Metastatic colonization is regarded the most rate- limiting step of metastasis and is subjected to regulation by a plethora of biological factors and processes. On one hand, regulation of metastatic colonization by autophagy appears to be stage- and context-dependent, whereas mechanistic characterization remains elusive. On the other hand, interactions between the tumor cells and their microenvironment in metastasis have long been appreciated, whether the secretome of tumor cells can effectively reshape the tumor microenvironment has not been elucidated mechanistically. In the present study, we have identified “SEC23A-S1008-BECLIN1-autophagy axis” in the autophagic regulation of metastatic colonization step, a mechanism that tumor cells can exploit autophagy to exert self-restrain for clonogenic proliferation before the favorable tumor microenvironment is established. Specifically, we employed a paired lung-derived oligometastatic cell line (OL) and the homologous polymetastatic cell line (POL) from human melanoma cell line M14 that differ in colonization efficiency. We show that S100A8 transported by SEC23A inhibits metastatic colonization via autocrine activation of autophagy. Furthermore, we verified the clinical relevance of our experimental findings by bioinformatics analysis of the expression of Sec23a and S100A8 and the clinical- pathological associations. We demonstrate that higher Sec23a and Atg5 expression levels appear to be protective factors and favorable diagnostic (TNM staging) and prognostic (overall survival) markers for skin cutaneous 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; melanoma (SKCM) and colon adenocarcinoma (COAD) patients. -

1714 Gene Comprehensive Cancer Panel Enriched for Clinically Actionable Genes with Additional Biologically Relevant Genes 400-500X Average Coverage on Tumor
xO GENE PANEL 1714 gene comprehensive cancer panel enriched for clinically actionable genes with additional biologically relevant genes 400-500x average coverage on tumor Genes A-C Genes D-F Genes G-I Genes J-L AATK ATAD2B BTG1 CDH7 CREM DACH1 EPHA1 FES G6PC3 HGF IL18RAP JADE1 LMO1 ABCA1 ATF1 BTG2 CDK1 CRHR1 DACH2 EPHA2 FEV G6PD HIF1A IL1R1 JAK1 LMO2 ABCB1 ATM BTG3 CDK10 CRK DAXX EPHA3 FGF1 GAB1 HIF1AN IL1R2 JAK2 LMO7 ABCB11 ATR BTK CDK11A CRKL DBH EPHA4 FGF10 GAB2 HIST1H1E IL1RAP JAK3 LMTK2 ABCB4 ATRX BTRC CDK11B CRLF2 DCC EPHA5 FGF11 GABPA HIST1H3B IL20RA JARID2 LMTK3 ABCC1 AURKA BUB1 CDK12 CRTC1 DCUN1D1 EPHA6 FGF12 GALNT12 HIST1H4E IL20RB JAZF1 LPHN2 ABCC2 AURKB BUB1B CDK13 CRTC2 DCUN1D2 EPHA7 FGF13 GATA1 HLA-A IL21R JMJD1C LPHN3 ABCG1 AURKC BUB3 CDK14 CRTC3 DDB2 EPHA8 FGF14 GATA2 HLA-B IL22RA1 JMJD4 LPP ABCG2 AXIN1 C11orf30 CDK15 CSF1 DDIT3 EPHB1 FGF16 GATA3 HLF IL22RA2 JMJD6 LRP1B ABI1 AXIN2 CACNA1C CDK16 CSF1R DDR1 EPHB2 FGF17 GATA5 HLTF IL23R JMJD7 LRP5 ABL1 AXL CACNA1S CDK17 CSF2RA DDR2 EPHB3 FGF18 GATA6 HMGA1 IL2RA JMJD8 LRP6 ABL2 B2M CACNB2 CDK18 CSF2RB DDX3X EPHB4 FGF19 GDNF HMGA2 IL2RB JUN LRRK2 ACE BABAM1 CADM2 CDK19 CSF3R DDX5 EPHB6 FGF2 GFI1 HMGCR IL2RG JUNB LSM1 ACSL6 BACH1 CALR CDK2 CSK DDX6 EPOR FGF20 GFI1B HNF1A IL3 JUND LTK ACTA2 BACH2 CAMTA1 CDK20 CSNK1D DEK ERBB2 FGF21 GFRA4 HNF1B IL3RA JUP LYL1 ACTC1 BAG4 CAPRIN2 CDK3 CSNK1E DHFR ERBB3 FGF22 GGCX HNRNPA3 IL4R KAT2A LYN ACVR1 BAI3 CARD10 CDK4 CTCF DHH ERBB4 FGF23 GHR HOXA10 IL5RA KAT2B LZTR1 ACVR1B BAP1 CARD11 CDK5 CTCFL DIAPH1 ERCC1 FGF3 GID4 HOXA11 IL6R KAT5 ACVR2A -
Type of the Paper (Article
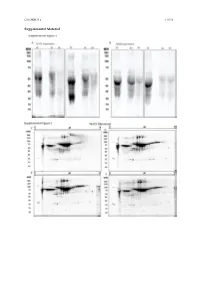
Cells 2020, 9, x 1 of 19 Supplemental Material Cells 2020, 9, x 2 of 19 Figure S1. Secretome enrichment: protocol optimization. 1D SDS-PAGE documentation of washing steps: Culture medium was substituted with FCS-free medium, which was changed every 2 h. The supernatants were then collected, and the proteins isolated and separated in 1D SDS-PAGE ((A) TK173 and (B) TK188). Proteins were stained with Flamingo fluorescent gel stain. Two-dimensional pattern of the proteins isolated from supernatant of TK173, (C) 2 h, (D) 4 h, (E) 6 h, and (F) 8 h after changing to FCS-free medium. (G) Cell secretome collected 24 h after elimination of the contaminating FCS-proteins with different washing steps. Proteins were stained with Flamingo fluorescent gel stain. Cells 2020, 9, x 3 of 19 Figure S2. 2-DE reference maps of secretomes; 150 μg proteins were loaded on an 11 cm IPG strip with a linear pH gradient PI 5–8 for IEF; 12% SDS-polyacrylamide gels were used for the second dimension. Proteins were stained with Flamingo fluorescent gel stain. Identified spots were assigned a number corresponding to that in their table. 2-DE maps from secretome of (A) TK173 control and (B) TGFβ1- treated ones. The 2-DE patterns revealed an alteration of secretome in stimulated TK173. Secretome patterns from TK173 treated with (C) ANG II and (D) PDGF. Cells 2020, 9, x 4 of 19 A Figure S3. Classification of the differentially expressed proteins upon ANG II, TGFβ1, or PDGF treatment in TK173. (A) Bar charts of the cellular component analyzed by STRAP biological function analysis in which the identified proteins from all treatments in both cell types are involved. -

Site-1 Protease Deficiency Causes Human Skeletal Dysplasia Due to Defective Inter-Organelle Protein Trafficking
Site-1 protease deficiency causes human skeletal dysplasia due to defective inter-organelle protein trafficking Yuji Kondo, … , Patrick M. Gaffney, Lijun Xia JCI Insight. 2018;3(14):e121596. https://doi.org/10.1172/jci.insight.121596. Research Article Cell biology Genetics Graphical abstract Find the latest version: https://jci.me/121596/pdf RESEARCH ARTICLE Site-1 protease deficiency causes human skeletal dysplasia due to defective inter-organelle protein trafficking Yuji Kondo,1 Jianxin Fu,1,2 Hua Wang,3 Christopher Hoover,1,4 J. Michael McDaniel,1 Richard Steet,5 Debabrata Patra,6 Jianhua Song,1 Laura Pollard,7 Sara Cathey,7 Tadayuki Yago,1 Graham Wiley,8 Susan Macwana,8 Joel Guthridge,8 Samuel McGee,1 Shibo Li,3 Courtney Griffin,1 Koichi Furukawa,9 Judith A. James,8 Changgeng Ruan,2 Rodger P. McEver,1,4 Klaas J. Wierenga,3 Patrick M. Gaffney,8 and Lijun Xia1,2,4 1Cardiovascular Biology Research Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma, USA. 2Jiangsu Institute of Hematology, MOH Key Laboratory of Thrombosis and Hemostasis, Collaborative Innovation Center of Hematology, The First Affiliated Hospital of Soochow University, Suzhou, China. 3Department of Pediatrics and 4Department of Biochemistry and Molecular Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma, USA. 5Complex Carbohydrate Research Center, University of Georgia, Georgia, Athens, USA. 6Department of Orthopaedic Surgery, Washington University School of Medicine, St. Louis, Missouri, USA. 7Greenwood Genetic Center, Greenwood, South Carolina, USA. 8Division of Genomics and Data Sciences, Arthritis and Clinical Immunology Program, Oklahoma Medical Research Foundation, Oklahoma City, Oklahoma, USA. 9Department of Biochemistry II, Nagoya University Graduate School of Medicine, Nagoya, Japan. -

Identification of Motifs in Cholera Toxin A1 Polypeptide That Are Required for Its Interaction with Human ADP-Ribosylation Factor 6 in a Bacterial Two-Hybrid System
Identification of motifs in cholera toxin A1 polypeptide that are required for its interaction with human ADP-ribosylation factor 6 in a bacterial two-hybrid system Michael G. Jobling and Randall K. Holmes* Department of Microbiology, University of Colorado Health Sciences Center, Denver, CO 80220 Edited by John J. Mekalanos, Harvard Medical School, Boston, MA, and approved October 20, 2000 (received for review September 14, 2000) The latent ADP-ribosyltransferase activity of cholera toxin (CT) that teins called ADP-ribosylation factors (ARFs; refs. 5 and 6). is activated after proteolytic nicking and reduction is associated ARFs are members of a highly conserved multigene family of with the CT A1 subunit (CTA1) polypeptide. This activity is stimu- small GTP-binding proteins, originally identified as activators of lated in vitro by interaction with eukaryotic proteins termed CT. They interact with several other proteins and are ubiqui- ADP-ribosylation factors (ARFs). We analyzed this interaction in a tously involved in membrane trafficking events (7). ARFs must modified bacterial two-hybrid system in which the T18 and T25 be in the GTP-bound form to be active. There are six mammalian fragments of the catalytic domain of Bordetella pertussis adenylate ARFs (five human) that fall into three classes: ARFs 1, 2, and cyclase were fused to CTA1 and human ARF6 polypeptides, respec- 3 (class I), ARFs 4 and 5 (class II), and ARF6 (class III). Class tively. Direct interaction between the CTA1 and ARF6 domains in I ARFs regulate the assembly of several types of vesicle coat these hybrid proteins reconstituted the adenylate cyclase activity complexes (8). -

ER-To-Golgi Trafficking and Its Implication in Neurological Diseases
cells Review ER-to-Golgi Trafficking and Its Implication in Neurological Diseases 1,2, 1,2 1,2, Bo Wang y, Katherine R. Stanford and Mondira Kundu * 1 Department of Pathology, St. Jude Children’s Research Hospital, Memphis, TN 38105, USA; [email protected] (B.W.); [email protected] (K.R.S.) 2 Department of Cell and Molecular Biology, St. Jude Children’s Research Hospital, Memphis, TN 38105, USA * Correspondence: [email protected]; Tel.: +1-901-595-6048 Present address: School of Life Sciences, Xiamen University, Xiamen 361102, China. y Received: 21 November 2019; Accepted: 7 February 2020; Published: 11 February 2020 Abstract: Membrane and secretory proteins are essential for almost every aspect of cellular function. These proteins are incorporated into ER-derived carriers and transported to the Golgi before being sorted for delivery to their final destination. Although ER-to-Golgi trafficking is highly conserved among eukaryotes, several layers of complexity have been added to meet the increased demands of complex cell types in metazoans. The specialized morphology of neurons and the necessity for precise spatiotemporal control over membrane and secretory protein localization and function make them particularly vulnerable to defects in trafficking. This review summarizes the general mechanisms involved in ER-to-Golgi trafficking and highlights mutations in genes affecting this process, which are associated with neurological diseases in humans. Keywords: COPII trafficking; endoplasmic reticulum; Golgi apparatus; neurological disease 1. Overview Approximately one-third of all proteins encoded by the mammalian genome are exported from the endoplasmic reticulum (ER) and transported to the Golgi apparatus, where they are sorted for delivery to their final destination in membrane compartments or secretory vesicles [1]. -

Figure S1. HAEC ROS Production and ML090 NOX5-Inhibition
Figure S1. HAEC ROS production and ML090 NOX5-inhibition. (a) Extracellular H2O2 production in HAEC treated with ML090 at different concentrations and 24 h after being infected with GFP and NOX5-β adenoviruses (MOI 100). **p< 0.01, and ****p< 0.0001 vs control NOX5-β-infected cells (ML090, 0 nM). Results expressed as mean ± SEM. Fold increase vs GFP-infected cells with 0 nM of ML090. n= 6. (b) NOX5-β overexpression and DHE oxidation in HAEC. Representative images from three experiments are shown. Intracellular superoxide anion production of HAEC 24 h after infection with GFP and NOX5-β adenoviruses at different MOIs treated or not with ML090 (10 nM). MOI: Multiplicity of infection. Figure S2. Ontology analysis of HAEC infected with NOX5-β. Ontology analysis shows that the response to unfolded protein is the most relevant. Figure S3. UPR mRNA expression in heart of infarcted transgenic mice. n= 12-13. Results expressed as mean ± SEM. Table S1: Altered gene expression due to NOX5-β expression at 12 h (bold, highlighted in yellow). N12hvsG12h N18hvsG18h N24hvsG24h GeneName GeneDescription TranscriptID logFC p-value logFC p-value logFC p-value family with sequence similarity NM_052966 1.45 1.20E-17 2.44 3.27E-19 2.96 6.24E-21 FAM129A 129. member A DnaJ (Hsp40) homolog. NM_001130182 2.19 9.83E-20 2.94 2.90E-19 3.01 1.68E-19 DNAJA4 subfamily A. member 4 phorbol-12-myristate-13-acetate- NM_021127 0.93 1.84E-12 2.41 1.32E-17 2.69 1.43E-18 PMAIP1 induced protein 1 E2F7 E2F transcription factor 7 NM_203394 0.71 8.35E-11 2.20 2.21E-17 2.48 1.84E-18 DnaJ (Hsp40) homolog. -

Lysophosphatidic Acid Activates Arf6 to Promote the Mesenchymal Malignancy of Renal Cancer
ARTICLE Received 6 Jun 2015 | Accepted 6 Jan 2016 | Published 8 Feb 2016 DOI: 10.1038/ncomms10656 OPEN Lysophosphatidic acid activates Arf6 to promote the mesenchymal malignancy of renal cancer Shigeru Hashimoto1,*, Shuji Mikami2,*,w, Hirokazu Sugino1, Ayumu Yoshikawa1, Ari Hashimoto1, Yasuhito Onodera1, Shotaro Furukawa1,3, Haruka Handa1, Tsukasa Oikawa1, Yasunori Okada4, Mototsugu Oya5 & Hisataka Sabe1 Acquisition of mesenchymal properties by cancer cells is critical for their malignant behaviour, but regulators of the mesenchymal molecular machinery and how it is activated remain elusive. Here we show that clear cell renal cell carcinomas (ccRCCs) frequently utilize the Arf6-based mesenchymal pathway to promote invasion and metastasis, similar to breast cancers. In breast cancer cells, ligand-activated receptor tyrosine kinases employ GEP100 to activate Arf6, which then recruits AMAP1; and AMAP1 then binds to the mesenchymal- specific protein EPB41L5, which promotes epithelial–mesenchymal transition and focal adhesion dynamics. In renal cancer cells, lysophosphatidic acid (LPA) activates Arf6 via its G-protein-coupled receptors, in which GTP-Ga12 binds to EFA6. The Arf6-based pathway may also contribute to drug resistance. Our results identify a specific mesenchymal molecular machinery of primary ccRCCs, which is triggered by a product of autotaxin and it is associated with poor outcome of patients. 1 Department of Molecular Biology, Graduate School of Medicine, Hokkaido University, Sapporo 060-8638, Japan. 2 Division of Diagnostic Pathology, Keio University Hospital, Tokyo 160-0016, Japan. 3 Department of Gastroenterological Surgery II, Graduate School of Medicine, Hokkaido University, Sapporo 060-8638, Japan. 4 Department of Pathology, Keio University School of Medicine, Tokyo 160-0016, Japan.