Identification of a Hypoxia-Related Lncrna Signature for the Prognosis of Colorectal Cancer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

REVISIONS Suzhou Talesun Solar Technologies Co., Ltd
REVISIONS SuZhou Talesun Solar Technologies Co., Ltd. REV ECN / NPA DESCRIPTION OF CHANGE CHK’D/DATE APP’D/DATE TITLE: LIMITED WARRANTY CERTIFICATE FOR TALESUN A0 07-2020 New Edition Caiping.huang Zhenzhou.gao 07-2020 07-2020 DOUBLE GLASS PHOTOVOLTAIC MODULE SPEC. NO.: PART NO.: TS-ET-125 N/A DRAWN BY: REV: Yujie.qian A0 SHEET 1 OF 1 TECHNICAL SPECIFICATION TITLE: LIMITED WARRANTY CERTIFICATE FOR SPEC. NO.: TS-ET-125 TALESUN DOUBLE GLASS PHOTOVOLTAIC REVISION: A0 MODULE EFFECTIVE DATE: 07-2020 Page 1 of 8 SuZhou Talesun Solar Technologies Co., Ltd. (“TALESUN”), hereby provide the limited warranty as below(“Limited Warranty”) described below to the initial owner of such Modules (the “Buyer”) This Limited Warranty applies exclusively to the new modules purchased from Talesun TALESUN reserve the right to amend the terms of this Limited Warranty as needed from time to time. Modules are defined in this Limited Warranty as photovoltaic solar modules manufactured by TALESUN or its authorized manufacturers, legitimately bearing “Talesun” brand, that are of the following product types: TD654P-XXX/TD654P-XXXN;TD660P-XXX/TD660P-XXXN;TD660M-XXX/TD660M-XXXN; TD672P-XXX/TD672P-XXXN;TD672M-XXX/TD672M-XXXN;TD6D60M-XXX;TD6D72M- XXX;TD6E60M-XXX/TD6E60M-XXXN ;TD6E72M-XXX/TD6E72M-XXXN ;TD6G72M-XXX ; TD6G60M-XXX;TDI72M-XXX;TD6I60M-XXX Note: M-Mono Module, P-Poly Module, The first D stands for double glass The second D-N type bifacial module E-P type bifacial module G-stand for half cutting module(9bb bifacial) I- stand for half cutting module(166 9bb bifacial) N-stand for none frame,The product model includes but is not limited to the above model and shall be subject to the specific model purchased by the customer. -

SGS-Safeguards 04910- Minimum Wages Increased in Jiangsu -EN-10
SAFEGUARDS SGS CONSUMER TESTING SERVICES CORPORATE SOCIAL RESPONSIILITY SOLUTIONS NO. 049/10 MARCH 2010 MINIMUM WAGES INCREASED IN JIANGSU Jiangsu becomes the first province to raise minimum wages in China in 2010, with an average increase of over 12% effective from 1 February 2010. Since 2008, many local governments have deferred the plan of adjusting minimum wages due to the financial crisis. As economic results are improving, the government of Jiangsu Province has decided to raise the minimum wages. On January 23, 2010, the Department of Human Resources and Social Security of Jiangsu Province declared that the minimum wages in Jiangsu Province would be increased from February 1, 2010 according to Interim Provisions on Minimum Wages of Enterprises in Jiangsu Province and Minimum Wages Standard issued by the central government. Adjustment of minimum wages in Jiangsu Province The minimum wages do not include: Adjusted minimum wages: • Overtime payment; • Monthly minimum wages: • Allowances given for the Areas under the first category (please refer to the table on next page): middle shift, night shift, and 960 yuan/month; work in particular environments Areas under the second category: 790 yuan/month; such as high or low Areas under the third category: 670 yuan/month temperature, underground • Hourly minimum wages: operations, toxicity and other Areas under the first category: 7.8 yuan/hour; potentially harmful Areas under the second category: 6.4 yuan/hour; environments; Areas under the third category: 5.4 yuan/hour. • The welfare prescribed in the laws and regulations. CORPORATE SOCIAL RESPONSIILITY SOLUTIONS NO. 049/10 MARCH 2010 P.2 Hourly minimum wages are calculated on the basis of the announced monthly minimum wages, taking into account: • The basic pension insurance premiums and the basic medical insurance premiums that shall be paid by the employers. -

News 20130307.Pdf
March 7, 2013 Mizuho Corporate Bank, Ltd. FOR GENERAL RELEASE Business Cooperation Agreements with the Administrative Committee of Changshu High Tech Industrial Park and the Zhangjiagang Free Trade Zone in Jiangsu Province, China Mizuho Corporate Bank, Ltd. (Yasuhiro Sato, President and CEO; MHCB) and Mizuho Corporate Bank (China), Ltd. (MHCB (China)) signed business cooperation agreements with the Administrative Committee of Changshu High Tech Industrial Park, CEDZ and the Zhangjiagang Free Trade Zone in Jiangsu Province in the People's Republic of China on March 6, 2013. The Administrative Committee of Changshu High Tech Industrial Park is located in the northern part of Suzhou city. It is a hub for high-tech manufacturing and Japanese corporations including high-tech-plant- related, precision instruments, and automotive components manufacturers have expanded their businesses into the area. The Zhangjiagang Free Trade Zone is a national-level free trade zone that is also located in the northern part of Suzhou city. Japanese corporations including iron and fine chemical manufacturers have been operating in the area for many years and more recently it is attracting industries such as automotive components and logistics. These business cooperation agreements aim for mutual cooperation toward economic development in the region by supporting Japanese corporations expanding their businesses there. As well as further progressing with advice on attracting industry to Changshu and Zhangjiagang cities in the Jiangsu area and cooperating in holding investment seminars, they will also strengthen support for Japanese corporations doing business in each of the areas. MHCB (China) used the opening of its Suzhou Branch in November 2010 as an opportunity to strengthen cooperation with economic development zones in the Suzhou area. -

Results Announcement for the Year Ended December 31, 2020
(GDR under the symbol "HTSC") RESULTS ANNOUNCEMENT FOR THE YEAR ENDED DECEMBER 31, 2020 The Board of Huatai Securities Co., Ltd. (the "Company") hereby announces the audited results of the Company and its subsidiaries for the year ended December 31, 2020. This announcement contains the full text of the annual results announcement of the Company for 2020. PUBLICATION OF THE ANNUAL RESULTS ANNOUNCEMENT AND THE ANNUAL REPORT This results announcement of the Company will be available on the website of London Stock Exchange (www.londonstockexchange.com), the website of National Storage Mechanism (data.fca.org.uk/#/nsm/nationalstoragemechanism), and the website of the Company (www.htsc.com.cn), respectively. The annual report of the Company for 2020 will be available on the website of London Stock Exchange (www.londonstockexchange.com), the website of the National Storage Mechanism (data.fca.org.uk/#/nsm/nationalstoragemechanism) and the website of the Company in due course on or before April 30, 2021. DEFINITIONS Unless the context otherwise requires, capitalized terms used in this announcement shall have the same meanings as those defined in the section headed “Definitions” in the annual report of the Company for 2020 as set out in this announcement. By order of the Board Zhang Hui Joint Company Secretary Jiangsu, the PRC, March 23, 2021 CONTENTS Important Notice ........................................................... 3 Definitions ............................................................... 6 CEO’s Letter .............................................................. 11 Company Profile ........................................................... 15 Summary of the Company’s Business ........................................... 27 Management Discussion and Analysis and Report of the Board ....................... 40 Major Events.............................................................. 112 Changes in Ordinary Shares and Shareholders .................................... 149 Directors, Supervisors, Senior Management and Staff.............................. -

Tier 1 Manufacturing Sites
TIER 1 MANUFACTURING SITES - Produced January 2021 SUPPLIER NAME MANUFACTURING SITE NAME ADDRESS PRODUCT TYPE No of EMPLOYEES Albania Calzaturificio Maritan Spa George & Alex 4 Street Of Shijak Durres Apparel 100 - 500 Calzificio Eire Srl Italstyle Shpk Kombinati Tekstileve 5000 Berat Apparel 100 - 500 Extreme Sa Extreme Korca Bul 6 Deshmoret L7Nr 1 Korce Apparel 100 - 500 Bangladesh Acs Textiles (Bangladesh) Ltd Acs Textiles & Towel (Bangladesh) Tetlabo Ward 3 Parabo Narayangonj Rupgonj 1460 Home 1000 - PLUS Akh Eco Apparels Ltd Akh Eco Apparels Ltd 495 Balitha Shah Belishwer Dhamrai Dhaka 1800 Apparel 1000 - PLUS Albion Apparel Group Ltd Thianis Apparels Ltd Unit Fs Fb3 Road No2 Cepz Chittagong Apparel 1000 - PLUS Asmara International Ltd Artistic Design Ltd 232 233 Narasinghpur Savar Dhaka Ashulia Apparel 1000 - PLUS Asmara International Ltd Hameem - Creative Wash (Laundry) Nishat Nagar Tongi Gazipur Apparel 1000 - PLUS Aykroyd & Sons Ltd Taqwa Fabrics Ltd Kewa Boherarchala Gila Beradeed Sreepur Gazipur Apparel 500 - 1000 Bespoke By Ges Unip Lda Panasia Clothing Ltd Aziz Chowdhury Complex 2 Vogra Joydebpur Gazipur Apparel 1000 - PLUS Bm Fashions (Uk) Ltd Amantex Limited Boiragirchala Sreepur Gazipur Apparel 1000 - PLUS Bm Fashions (Uk) Ltd Asrotex Ltd Betjuri Naun Bazar Sreepur Gazipur Apparel 500 - 1000 Bm Fashions (Uk) Ltd Metro Knitting & Dyeing Mills Ltd (Factory-02) Charabag Ashulia Savar Dhaka Apparel 1000 - PLUS Bm Fashions (Uk) Ltd Tanzila Textile Ltd Baroipara Ashulia Savar Dhaka Apparel 1000 - PLUS Bm Fashions (Uk) Ltd Taqwa -
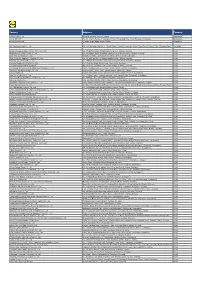
Factory Address Country
Factory Address Country Durable Plastic Ltd. Mulgaon, Kaligonj, Gazipur, Dhaka Bangladesh Lhotse (BD) Ltd. Plot No. 60&61, Sector -3, Karnaphuli Export Processing Zone, North Potenga, Chittagong Bangladesh Bengal Plastics Ltd. Yearpur, Zirabo Bazar, Savar, Dhaka Bangladesh ASF Sporting Goods Co., Ltd. Km 38.5, National Road No. 3, Thlork Village, Chonrok Commune, Korng Pisey District, Konrrg Pisey, Kampong Speu Cambodia Ningbo Zhongyuan Alljoy Fishing Tackle Co., Ltd. No. 416 Binhai Road, Hangzhou Bay New Zone, Ningbo, Zhejiang China Ningbo Energy Power Tools Co., Ltd. No. 50 Dongbei Road, Dongqiao Industrial Zone, Haishu District, Ningbo, Zhejiang China Junhe Pumps Holding Co., Ltd. Wanzhong Villiage, Jishigang Town, Haishu District, Ningbo, Zhejiang China Skybest Electric Appliance (Suzhou) Co., Ltd. No. 18 Hua Hong Street, Suzhou Industrial Park, Suzhou, Jiangsu China Zhejiang Safun Industrial Co., Ltd. No. 7 Mingyuannan Road, Economic Development Zone, Yongkang, Zhejiang China Zhejiang Dingxin Arts&Crafts Co., Ltd. No. 21 Linxian Road, Baishuiyang Town, Linhai, Zhejiang China Zhejiang Natural Outdoor Goods Inc. Xiacao Village, Pingqiao Town, Tiantai County, Taizhou, Zhejiang China Guangdong Xinbao Electrical Appliances Holdings Co., Ltd. South Zhenghe Road, Leliu Town, Shunde District, Foshan, Guangdong China Yangzhou Juli Sports Articles Co., Ltd. Fudong Village, Xiaoji Town, Jiangdu District, Yangzhou, Jiangsu China Eyarn Lighting Ltd. Yaying Gang, Shixi Village, Shishan Town, Nanhai District, Foshan, Guangdong China Lipan Gift & Lighting Co., Ltd. No. 2 Guliao Road 3, Science Industrial Zone, Tangxia Town, Dongguan, Guangdong China Zhan Jiang Kang Nian Rubber Product Co., Ltd. No. 85 Middle Shen Chuan Road, Zhanjiang, Guangdong China Ansen Electronics Co. Ning Tau Administrative District, Qiao Tau Zhen, Dongguan, Guangdong China Changshu Tongrun Auto Accessory Co., Ltd. -

Factory List September 2020 Liste Der Fabriken September 2020 Liste Des
ListeListeFactory der des Fabriken usines List SeptembreSeptember 2020 D06 Factory List 2020.indd 1 15/09/2020 16:28 FactoryNameNom De Der NameL'Usine Fabrik FactoryAnschriftAdresse AddressDe Der L'Usine Fabrik CountryLandPays ProductProdukt-Catégorie NumberAnzahlNombre Der D'EmployésOf Workers % % CategorykategorieDe Produit Beschäftigten*Correct*Données As Mises Of À Jour En FemaleWeiblichFemelle MaleMännMâle - SeptemberJuin 2020 2020 *Zutreffend Mit Wirkung Ab lich September 2020 Afa 3 Calzature Sh.P.K Afa 3 Calzature Sh.p.k, Berat, Albania Albania WW 221 73 27 Grace Glory Garments Preykor Village, Lumhach Commune, Cambodia Mini 2483 90 10 Angsnoul District, Kandal Popmode Dyontex (Ningbo) No 72-106 Gongmao 1 Road, Ishigang China WW, Mini 492 68 32 Limited Industrial Zone, Ningbo, 315171 Zhucheng Tianyao Garment Zangkejia Road, Textile & Garment China Mini 306 79 21 Co. Ltd. Industrial Park, Longdu Sub-District, Zhucheng City, Weifang City, Shandong Province Fujian Tancome Apparel Baijin Industrial Park, Baizhong Town China MW, Mini 161 68 32 Minging Country, Fuzhou City Anhui Baode Clothing Co Economic Zone, Chengdong Town, China Mini 155 79 21 Ltd Yongqiao District, Suzhou City Dezhou Excellent Garment No 16, Geruide Road Dezhou Economic China MW, Mini 260 92 8 Distric, Dezhou City Shandong Province Goldenmine Co Shuanghu Village, Hengcun Town, Tonglu China Mini 44 64 36 County, Hangzhou City, Zhejiang Province CWG No. 2 Xie Wu First Industry Area, China Mini 19 58 42 Hengshan Village, Shipal Town, Guangdong Ningbo Fenghe No.135 Jinyuan Road, Part B, Economic China Mini 290 85 15 Development, Zhenhai, Ningbo, 315221, Taishan City Taicheng No 106, Qiaohu Road, Taicheng Town, China Mini 172 84 16 Together Garment Factory Taishan City, Guangdong, 529200 Shenzhen Fuhowe Fashion 1-3/F Building, 10 Nangang Industrial Park China WW 221 35 65 Co Ltd Phase 1, Xili, Nanshan District, Shenzhen, 518055 Auro (Xinfeng) Fashion Co No. -

Transmissibility of Hand, Foot, and Mouth Disease in 97 Counties of Jiangsu Province, China, 2015- 2020
Transmissibility of Hand, Foot, and Mouth Disease in 97 Counties of Jiangsu Province, China, 2015- 2020 Wei Zhang Xiamen University Jia Rui Xiamen University Xiaoqing Cheng Jiangsu Provincial Center for Disease Control and Prevention Bin Deng Xiamen University Hesong Zhang Xiamen University Lijing Huang Xiamen University Lexin Zhang Xiamen University Simiao Zuo Xiamen University Junru Li Xiamen University XingCheng Huang Xiamen University Yanhua Su Xiamen University Benhua Zhao Xiamen University Yan Niu Chinese Center for Disease Control and Prevention, Beijing City, People’s Republic of China Hongwei Li Xiamen University Jian-li Hu Jiangsu Provincial Center for Disease Control and Prevention Tianmu Chen ( [email protected] ) Page 1/30 Xiamen University Research Article Keywords: Hand foot mouth disease, Jiangsu Province, model, transmissibility, effective reproduction number Posted Date: July 30th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-752604/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 2/30 Abstract Background: Hand, foot, and mouth disease (HFMD) has been a serious disease burden in the Asia Pacic region represented by China, and the transmission characteristics of HFMD in regions haven’t been clear. This study calculated the transmissibility of HFMD at county levels in Jiangsu Province, China, analyzed the differences of transmissibility and explored the reasons. Methods: We built susceptible-exposed-infectious-asymptomatic-removed (SEIAR) model for seasonal characteristics of HFMD, estimated effective reproduction number (Reff) by tting the incidence of HFMD in 97 counties of Jiangsu Province from 2015 to 2020, compared incidence rate and transmissibility in different counties by non -parametric test, rapid cluster analysis and rank-sum ratio. -

LIMITED WARRANTY CERTIFICATE for TALESUN A0 07-2019 New Edition Zhenzhou.Gao Mike.Qiu 07-2019 07-2019 DOUBLE GLASS PHOTOVOLTAIC MODULE SPEC
REVISIONS SuZhou Talesun Solar Technologies Co., Ltd. REV ECN / NPA DESCRIPTION OF CHANGE CHK’D/DATE APP’D/DATE TITLE: LIMITED WARRANTY CERTIFICATE FOR TALESUN A0 07-2019 New Edition Zhenzhou.gao Mike.qiu 07-2019 07-2019 DOUBLE GLASS PHOTOVOLTAIC MODULE SPEC. NO.: PART NO.: TS-ET-073 N/A DRAWN BY: REV: Yujie.qian A0 SHEET 1 OF 1 TECHINICAL SPECIFICATION TITLE: LIMITED WARRANTY CERTIFICATE FOR SPEC. NO.: TS-ET-073 TALESUN DOUBLE GLASS PHOTOVOLTAIC REVISION: A0 MODULE EFFECTIVE DATE: 07-2019 Page 1 of 7 SuZhou Talesun Solar Technologies Co., Ltd. (“TALESUN”) warrants to the first customer installing (for its own use) (the “Buyer”) its “TALESUN” double glass photovoltaic modules as: 1. TD654P-XXX/TD654P-XXXN(XXX=210,215,220,225,230,235,240,245,250) 2. TD660P-XXX/TD660P-XXXN ( XXX=240,245,250,255,260,265,270,275,280,285 , 290,295,300,305,310,315,320,325,330) 3. TD660M-XXX/TD660M-XXXN ( XXX=270,275,280,285,290,295,300,305,310,310 315,320,325,330,335,340,345) 4. TD672P-XXX/TD672P-XXXN ( XXX=260,265,270,275,280,285,290,295,300,305 , 310,315,320,325,330,335,340,345,350,355,360,365,370,375,380,385) 5. TD672M-XXX/TD672M-XXXN ( XXX=325,330,335,340,345,350,355,360,365,370 , 375,380,385,390,395,400,405,410,415,420) 6. TD6D60M-XXX(XXX=300,305,310,315,320,325,330,335,340) 7. TD6D72M- XXX(XXX=350,355,340,345,350,355,360,365,370,375,380,385,390,395, 400,405,410) 8. -

1997 1200 5000 6 2,300 Changshu, Jiangsu CHINA 1997 600 10000 4
Zhejiang Eastern Haobo Pipes Co., Ltd Open Cut Pipe (SN5000 - SN20,000) - Projects within Peoples Republic of China YEAR DN in mm SN PN LENGTH PROJECT COUNTRY 1997 1200 5000 6 2,300 Changshu, Jiangsu CHINA 1997 600 10000 4 1,800 Sanming, Fujiang CHINA 1997 600 5000 6 4,000 Shangyu, Zhejiang CHINA 1997 600 10000 6 4,000 Oubei, Yongjia, Zhejiang CHINA 1998 1000/1400 7500 6 11,000 Cangnan, Wenzhou, Zhejiang CHINA 1998 1200/1400 10000 6 26,750 Caoyong, Wenzhou, Zhejiang CHINA 1998 800 7500 6 2,500 Oubei, Yongjia, Zhejiang CHINA 1998 1000/1400 5000 4 1,500 Wenzhou Economic&Tech. Development Zone CHINA 1998 600/800 5000 6 9,000 Changshu, Jiangsu CHINA 1998 500/600 5000 6 13,500 Shangyu, Zhejiang CHINA 1998 1400 5000 4 1,200 Xiamen, Fujian CHINA 1999 500 5000 6 11,000 Changshu, Jiangsu CHINA 1999 700 10000 10 3,000 Dalian Economic&Tech. Development Zone CHINA 1999 1200 5000 6 6,000 Fujian CHINA 1999 900 10000 7 4,400 Taizhou, Jiangsu CHINA 1999 1800 10000 4 240 Pudong, Shanghai CHINA 1999 1000 5000 4 120 Changshan,zhengjiang CHINA 1999 800/1000 10000 10 4,600 Tianjing Economic&Tech. Development Zone CHINA 1999 500/600 5000 6 4,600 Shangyu, Zhejiang CHINA 1999 600 10000 8 600 Wenzhou, Zhejiang CHINA 1999 600 5000 6 600 Woyang, Anhui CHINA 1999 1200 5000/10000 4 2,200 Shaoxing, Zhejiang CHINA 1999 500 10000 4 558 Longquan, Chengdu, Sichuan CHINA 1999 500 10000 6 650 Fengxian, Shanghai CHINA 1999 400/600 5000 6 12,000 Shangyu, Zhejiang CHINA 1999 1400 10000 6 7,000 Hangzhou Economic&Tech . -

The Information Contained in This Document Is Kindly Provided by BIMCO
The information contained in this document is kindly provided by BIMCO. Please notice we take no legal responsibility its accuracy. Changes to the preventive measure might apply with little to no notice. We advise BIMCO members to contact the secretariat for the latest available updates. Please find below contact details: Maritime Information: [email protected] Wayne Zhuang, Regional Manager, Asia: [email protected] Maite Klarup, General Manager, Singapore: [email protected] Elena Tassioula, General Manager, Greece and Cyprus: [email protected] Kindly notice we provide information on restrictions and port related matters as a complementary member benefit. Non-members are encouraged to contact BIMCO for more information on member benefits. Please find below contact details: Membership: [email protected] Erik Jensby, Head of Membership: [email protected] Contents Details on prevention measures by region ....................................................................................... 4 North-east China (Dalian, Dandong, Jinzhou, Yingkou) ................................................................ 4 Tianjin ........................................................................................................................................ 4 Ports in Hebei (Tangshan, Huanghua, Qinhuangdao) ................................................................... 5 Tangshan Port ............................................................................................................................ 5 Caofeidian Port ......................................................................................................................... -

Wuxi Apptec Co., Ltd.* 無錫藥明康德新藥開發股份有限公司
THIS CIRCULAR IS IMPORTANT AND REQUIRES YOUR IMMEDIATE ATTENTION If you are in any doubt as to any aspect of this circular or as to the action to be taken, you should consult a stockbroker or other registered dealer in securities, a bank manager, solicitor, professional accountant or other professional adviser. If you have sold or transferred all your shares in WuXi AppTec Co., Ltd.* (無錫藥明康德新藥開發股份有限公司), you should at once hand this circular, together with the enclosed form of proxy, to the purchaser or transferee or to the bank, stockbroker or other agent through whom the sale or transfer was effected for transmission to the purchaser or transferee. Hong Kong Exchanges and Clearing Limited and The Stock Exchange of Hong Kong Limited take no responsibility for the contents of this circular, make no representation as to its accuracy or completeness and expressly disclaim any liability whatsoever for any loss howsoever arising from or in reliance upon the whole or any part of the contents of this circular. WUXI APPTEC CO., LTD.* 無錫藥明康德新藥開發股份有限公司 (A joint stock company incorporated in the People’s Republic of China with limited liability) (Stock Code: 2359) (1) WORK REPORT OF THE BOARD OF DIRECTORS FOR THE YEAR 2019; (2) WORK REPORT OF THE SUPERVISORY COMMITTEE FOR THE YEAR 2019; (3) ANNUAL REPORT FOR THE YEAR 2019; (4) FINANCIAL REPORT FOR THE YEAR 2019; (5) PROPOSED 2019 PROFIT DISTRIBUTION PLAN; (6) PROPOSED RE-ELECTION OF DIRECTORS; (7) PROPOSED ELECTION OF EXECUTIVE DIRECTOR; (8) PROPOSED RE-ELECTION OF SHAREHOLDER REPRESENTATIVE SUPERVISORS;