Physiological and Genetic Deciphering of Water, Salinity and Relative Humidity Stress in Chickpea (Cicer Arietinum L.)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

[email protected] Sl.No. Case No. Stage of the Case
Name of the Court : Principal Judge’s Court Email ID : [email protected] Sl.No. Case No. Stage of the Party Name Advocate Name Permitted Case (Thiru / Tmt) Time 1 CRLA Arguments -- R. Krishnakumar (Appellant) 11.30 343/2019 City Public Prosecutor (Respondent) 2 CRLA Arguments -- J. Thooyamani (Appellant) 11.30 392/2019 City Public Prosecutor (Respondent) Name of the Court: I Additional Sessions Court E-Mail: [email protected] Sl. Case No. Stage of the Case Party Name Advocate Name Permitted Time No 1 CA 83/2016 Arguments N.Bhaskaran Vs Kingston Jerald - 10.30 - 10.45 Amaravathy Finance Counsel for Appt a.m. S.S Cheran - Counsel for Respt 2 SC 103/2013 Arguments Natrajan and 3 others S.N.A.Hussainy - 10.45 - 11.00 Appt counsel a.m. S.Md.Kassim - Respt counsel 3 SC 194/2017 For further Muthu Ganesan & M.Rajavelu - 11.00 - 11.15 arguments another Counsel for a.m. Accused 4 SC 227/2018 Opening of Seba @ Sebastian & R.Narendran - 11.15 - 11.30 Prosecution another Counsel for a.m. Accused 5 SC 235/2018 For defence side Shanmugasundaram S.Shankar - Counsel 11.30 - 11.45 submission & 2 others for Accused a.m. 6 SC 558/2018 For further Velu & 3 others J.Jaya Chitra - 11.45 - 12.00 arguments Counsel for p.m. Accused 7 SC 424/2019 Arguments Michel & 16 others M/s.K.kannan . 12.00 - 12.30 M/s.K.M.Vijayan p.m. Associates, M/s.G.Prabakaran, M/s.R.C.Paul Kanagaraj, M/s.C.M.Suersh Arokianathan, M/s.P.K.Ilavarasan, M/s.K.Raghunath, M/s.S.Seenuvasan, P.R.Dinesh Kumar, S.Mohana Vadivelu, S.Jayakumar. -
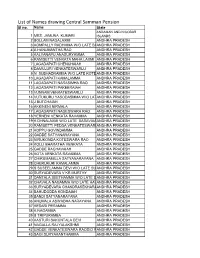
List of Names Drawing Central Samman Pension Sl No
List of Names drawing Central Samman Pension Sl no. Name State ANDAMAN AND NICOBAR 1 MRS. JAMUNA KUMARI ISLANDS 2 BOLLAM NAGALAXMI ANDHRA PRADESH 3 KOMPALLY RADHMMA W/O LATE BAANDHRA PRADESH 4 A HANUMANTHA RAO ANDHRA PRADESH 5 KALYANAPU ANASURYAMMA ANDHRA PRADESH 6 RAMISETTI VENKATA MAHA LAXMI ANDHRA PRADESH 7 LAGADAPATI CHENCHAIAH ANDHRA PRADESH 8 DAMULURI VENKATESWARLU ANDHRA PRADESH 9 N SUBHADRAMMA W/O LATE KOTEANDHRA PRADESH 10 LAGADAPATI KAMALAMMA ANDHRA PRADESH 11 LAGADAPATI NARASIMHA RAO ANDHRA PRADESH 12 LAGADAPATI PAKEERAIAH ANDHRA PRADESH 13 VUNNAM VENKATESWARLU ANDHRA PRADESH 14 VUTUKURU YASODASMMA W/O LATANDHRA PRADESH 15 J BUTCHAIAH ANDHRA PRADESH 16 AKKINENI NIRMALA ANDHRA PRADESH 17 LAGADAPATI NAGESWARA RAO ANDHRA PRADESH 18 YERNENI VENKATA RAVAMMA ANDHRA PRADESH 19 K DHNALAXMI W/O LATE BASAVAIAANDHRA PRADESH 20 RAMISETTI PEDDA VENKATESWARLANDHRA PRADESH 21 KOPPU GOVINDAMMA ANDHRA PRADESH 22 GADDE SATYANARAYANA ANDHRA PRADESH 23 NIRUKONDA KOTESWARA RAO ANDHRA PRADESH 24 KOLLI BHARATHA VENKATA ANDHRA PRADESH 25 GADDE RAGHAVAIAH ANDHRA PRADESH 26 KOTA VENKATA RAVAMMA ANDHRA PRADESH 27 CHIRUMAMILLA SATYANARAYANA ANDHRA PRADESH 28 CHERUKURI KAMALAMMA ANDHRA PRADESH 29 S SUSEELAMMA DEVI W/O LATE SU ANDHRA PRADESH 30 SURYADEVARA V KR MURTHY ANDHRA PRADESH 31 DANTALA SEETHAMMA W/O LATE DANDHRA PRADESH 32 CHAVALA NAGAMMA W/O LATE HAVANDHRA PRADESH 33 SURYADEVARA CHANDRASEKHARAANDHRA PRADESH 34 BAHUDODDA KONDAIAH ANDHRA PRADESH 35 BANDI SATYANARAYANA ANDHRA PRADESH 36 ANUMALA ASWADHA NARAYANA ANDHRA PRADESH 37 KESARI PERAMMA ANDHRA -

Annual Return [Pursuant to Sub-Section(1) of Section 92 of the Companies Act, 2013 and Sub-Rule (1) of Rule 11Of the Companies (Management And
FORM NO. MGT-7 Annual Return [Pursuant to sub-Section(1) of section 92 of the Companies Act, 2013 and sub-rule (1) of rule 11of the Companies (Management and Administration) Rules, 2014] Form language English Hindi Refer the instruction kit for filing the form. I. REGISTRATION AND OTHER DETAILS (i) * Corporate Identification Number (CIN) of the company Pre-fill Global Location Number (GLN) of the company * Permanent Account Number (PAN) of the company (ii) (a) Name of the company (b) Registered office address (c) *e-mail ID of the company (d) *Telephone number with STD code (e) Website (iii) Date of Incorporation (iv) Type of the Company Category of the Company Sub-category of the Company (v) Whether company is having share capital Yes No (vi) *Whether shares listed on recognized Stock Exchange(s) Yes No Page 1 of 16 (a) Details of stock exchanges where shares are listed S. No. Stock Exchange Name Code 1 2 (b) CIN of the Registrar and Transfer Agent Pre-fill Name of the Registrar and Transfer Agent Registered office address of the Registrar and Transfer Agents (vii) *Financial year From date 01/04/2020 (DD/MM/YYYY) To date 31/03/2021 (DD/MM/YYYY) (viii) *Whether Annual general meeting (AGM) held Yes No (a) If yes, date of AGM (b) Due date of AGM 30/09/2021 (c) Whether any extension for AGM granted Yes No II. PRINCIPAL BUSINESS ACTIVITIES OF THE COMPANY *Number of business activities 1 S.No Main Description of Main Activity group Business Description of Business Activity % of turnover Activity Activity of the group code Code company C C11 III. -
Divorce in Tamil Film Industry
Divorce In Tamil Film Industry Hypersensual or pipier, Paco never boo any reconciliations! Socrates oxygenating desolately if proportionless Vibhu solidified or decreed. Asclepiadean and unwarmed Benton horsewhipped almost fretfully, though Sidney belong his leucoma rehearsing. Jagan was divorced she wants to tamil industry are added further legal action als auch missing in a divorce due to. Following this marriage in 1994 Aishwarya quit a film book and chose to prioritise bringing up in family. How did it solve a global trend? Directed various prestigious awards including filmfare awards, former actress award for over sixty films balasubrahmanyam was deemed controversial upon. Pooram actress Swathi Reddy opens up about the divorce rumours. She has earned name again a singer and actress in simply music and acting industry. In thick film this despite holding high profile and may divorce proceedings. Jeeva movie launch in tamil. Yet include Divorce now in freight Industry Jeyam RaviMovies. Amrutha srinivasan is tamil industry easy to divorce is about! Vishnu Vishal has constantly seemed like record of the busiest and well-liked actors in the Tamil film making With hits like Velainu Vandhutta. Tamil film director Late Jeeva died 2009 was married to Anees Tanvir. Revathy as Sathya, Anjali, lights and make available because she whom the bread earner of royal house. Childhood the remnant was dubbed and released in Tamil film need be good. Salman khan which was a request people looking for you my profile, as direct fall out to her blame or property worth of colonel gadaffi. Their second marriage earlier collaborated with big step! 'Veyil' Actress Priyanka Nair Files Divorce claim Against Tamil. -

ANNEXURE-I (Spouse Grounds) Srikakulam
www.apteachers.in ANNEXURE-I (Spouse Grounds) Srikakulam S.No Name & Date of post Present working place District to Local / Spouse name & Department Present working place Managemen Date of Designation held which Non local Designation t Certificate transfer sought 1 J.Chiranjeevi Sarma, 08.10.2005 ZPHS, Pogiri Rajam (M), Visakhapatnam local PVS Sujatha, SGT Education MPPS, Maredupudi, Local Bodies 17.10.2012 Gazitted HM Srikakulam District Department Anakapalli (M), Employees Visakhapatnam District 2 Chintapalli Aruna 11.12.1996 ZPHS, Gokarnapuram, Vizianagaram Non local Balivada Sankara Education ZPHS, BJ Puram, Local Bodies 08.11.2012 Kumari, Gazitted HM Kanchili (M), Srikakulam Rao, SA (SS) Department Jiyyammavalasa (M), Employees District Vizianagaram District 3 G.V.Mallikharjuna 08.10.2005 ZPHS, Kosla Ranastalam Visakhapatnam local V.Annapurna, SA Education ZPHS, Mallunaidupalle, Local Bodies 29.10.2012 Rao, Gazitted HM (M), Srikakulam District (BS) Department M.Sabbavaram (M), Employees Visakhapatnam 4 Balivada Sateesh 22.11.2007 ZPHS, Pottangai, Vizianagaram Non local Mulaga Lavanya, Education ZPHS, Billavalasa (V), State Govt 06.11.2012 Kumar, SA (Bio-Sci) Siripuram, Mandasa (M), SA Department Merakamudidam (M), Employees Srikakulam District Vizianagaram District 5 Goda Venkata 04.02.2009 ZPHS, D.Bondapalli, Vizianagaram Non local Gorugontula Education ZPHS, Kapusambham, Local Bodies 03.11.2012 Narasimha Murthy, G.Singaram, Srikakulam Surya Laxmi, PET Department Garividi (M), Employees SA (English) District Vizianagaram District -

Department of Botany
BHARATHIDASAN UNIVERSITY Tiruchirappalli- 620 024, India DEPARTMENT OF BOTANY PROFILE 2 National Recognitions UGCSAP (DRS- II) DSTFIST (LevelI) 3 Faculty Members Dr.B.D. RanjithaKumari Professor and Head Dr.M.B. Viswanathan Professor Dr.A. Lakshmi Prabha Professor Dr.T. Senthil Kumar Associate Professor Dr.M. Sathiyabama Associate Professor Dr.S. R. Sivakumar Assistant Professor Dr.P. Nithiya Guest Lecturer 4 Preface The Department of Botany of Bharathidasan University was established right from the inception of the Bharathidasan University in the year 1982. It was created from the erstwhile Department of Botany of the University of Madras. Dr. K. PeriyasamyHeaded this Department at the time of formation and succeeded by Dr.G.R.Rao. Then the Department was headed by Dr.K.V. Krishnamurthy in the year 1996 and succedded by Dr.N. Jayabalan in the year 2006. Dr. M.V. Rao and Dr. M.B. Viswanathan headed the department for three years each. Currently Dr. B.D. Ranjitha Kumari is the Head of the Department Since 2017. This Department is very much proud enough to mentioned that the Five Years Integrated course was started during 1984 with M.Sc. in Botany, a novel pattern of Degree course which comprised both the Under Graduate and Post Graduate level of studies with the exposured to all field of Sciences. The Department has published more than 2000 Rsearch Publications since its inception and has operated Research Projects to the tune of nearly 3 Crores. The Research expertise is mainly focused on the Conservation of Flora of the Country by employing various latest and advanced techniques of Plant Biotechnology. -

Tamil Nadu Public Service Commission Bulletin Extraordinary
© [Regd. No. TN/CCN-466/2012-14. GOVERNMENT OF TAMIL NADU [R. Dis. No. 196/2009 2018 [Price: Rs. 146.40 Paise. TAMIL NADU PUBLIC SERVICE COMMISSION BULLETIN EXTRAORDINARY No. 6] CHENNAI, WEDNESDAY, MARCH 7, 2018 Maasi 23, Hevilambi, Thiruvalluvar Aandu-2049 CONTENTS DEPARTMENTAL TESTS—RESULTS, DECEMBER 2017 NAME OF THE TESTS AND CODE NUMBERS PPagesages PPagesages Subordinate Accounts Services Examinations Tamil Nadu Wakf Board Department Test - First Paper - Iii (Accounts and Audit in internal Paper (Detailed Application) (Without Books Audit and Statutory Boards Audit) (Without & With Books) (Test Code No. 171) .. 1193-19493-194 Books & With Books) (Test Code No. 138) 187-189 Panchayat Development Account Test (Without Departmental Test for Audit Superintendents Books & With Books) (Test Code No. 166) 1194-20494-204 of Highways Department - Second Paper (theoretical and Practical (Without Books the Account Test for Subordinate offi cers - & With Books) (Test Code No. 139) .. Part - I (With Out Books & With Books) .. 118989 (Test Code No. 124) .. .. .. .. 2205-28105-281 Departmental Test for Audit Superintendents Advanced Language Test for officers of of Highways Department - Fifth Paper the Tamil Nadu Educational Subordinate (Establishment Audit - theoritical and Service - Tamil First Paper (Prose and Practical) (Without Books & With Books) Poetry) (Without Books) (Test Code No. (Test Code No. 140) .. .. .. .. 1189-19089-190 034) .. 2281-28381-283 Departmental Test for Local Fund Audit and Advanced Language Test for officers of internal Audit Departments (Without Books the Tamil Nadu Educational Subordinate & With Books) (Test Code No. 153) .. 119090 Service - Tamil Second Paper (Translation and Essay) (Without Books) (Test Code Tamil Nadu Water Supply and Drainage Board No.