DIFFUSION MAPS 43 5.1 Clustering
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Anastasios Kyrillidis
Anastasios Kyrillidis CONTACT 3119 Duncan Hall INFORMATION 6100 Main Street, 77005 Tel: (+1) 713-348-4741 Houston, United States E-mail: [email protected] Website: akyrillidis.github.io RESEARCH Optimization for machine learning, convex and non-convex analysis and optimization, structured low dimensional INTERESTS models, large-scale computing, quantum computing. CADEMIC Rice University, Houston, USAA APPOINTMENTS Noah Harding Assistant Professor at Computer Science Dept. July 2018 - now University of Texas at Austin, Austin, USA Simons Foundation Postdoctoral Researcher November 2014 - August 2017 Member of WNCG PROFESSIONAL IBM T.J. Watson Research Center, New York (USA) APPOINTMENTS Goldstine PostDoctoral Fellow September 2017 - July 2018 EDUCATION Ecole´ Polytechnique F´ed´eralede Lausanne (EPFL), Lausanne, Switzerland Ph.D., School of Computer and Communication Sciences, September 2010 - October 2014. TEACHING AND Instructor SUPERVISING Rice University EXPERIENCE • COMP 414/514 — Optimization: Algorithms, complexity & approximations — Fall ‘19 • Students enrolled: 30 (28 G/2 UG) • Class evaluation — Overall quality: 1.54 (Rice mean: 1.76); Organization: 1.57 (Rice mean: 1.75); Chal- lenge: 1.5 (Rice 1.73) • Instructor evaluation — Effectiveness: 1.43 (Rice mean: 1.67); Presentation: 1.39 (Rice mean: 1.73); Knowledge: 1.29 (Rice mean: 1.56). — Fall ‘20 • Students enrolled: 30 (8 G/22 UG) • Class evaluation — Overall quality: 1.29 (Rice mean: 1.69); Organization: 1.28 (Rice mean: 1.69); Chal- lenge: 1.31 (Rice 1.68) • Instructor evaluation — Effectiveness: 1.21 (Rice mean: 1.58); Presentation: 1.17 (Rice mean: 1.63); Knowledge: 1.14 (Rice mean: 1.49). • COMP 545 — Advanced topics in optimization: From simple to complex ML systems — Spring ‘19 • Students enrolled: 10 • Class evaluation — Overall quality: 1.31 (Rice mean: 1.78); Organization: 1.23 (Rice mean: 1.79); Chal- lenge: 1.46 (Rice mean: 1.75). -

University of California San Diego
UNIVERSITY OF CALIFORNIA SAN DIEGO Sparse Recovery and Representation Learning A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Mathematics by Jingwen Liang Committee in charge: Professor Rayan Saab, Chair Professor Jelena Bradic Professor Massimo Franceschetti Professor Philip E. Gill Professor Tara Javidi 2020 Copyright Jingwen Liang, 2020 All rights reserved. The dissertation of Jingwen Liang is approved, and it is ac- ceptable in quality and form for publication on microfilm and electronically: Chair University of California San Diego 2020 iii DEDICATION To my loving family, friends and my advisor. iv TABLE OF CONTENTS Signature Page . iii Dedication . iv Table of Contents . .v List of Figures . vii Acknowledgements . viii Vita .............................................x Abstract of the Dissertation . xi Chapter 1 Introduction and Background . .1 1.1 Compressed Sensing and low-rank matrix recovery with prior infor- mations . .1 1.2 Learning Dictionary with Fast Transforms . .3 1.3 Deep Generative Model Using Representation Learning Techniques5 1.4 Contributions . .6 Chapter 2 Signal Recovery with Prior Information . .8 2.1 Introduction . .8 2.1.1 Compressive Sensing . .9 2.1.2 Low-rank matrix recovery . 10 2.1.3 Prior Information for Compressive Sensing and Low-rank matrix recovery . 12 2.1.4 Related Work . 13 2.1.5 Contributions . 21 2.1.6 Overview . 22 2.2 Low-rank Matrices Recovery . 22 2.2.1 Problem Setting and Notation . 22 2.2.2 Null Space Property of Low-rank Matrix Recovery . 23 2.3 Low-rank Matrix Recovery with Prior Information . 31 2.3.1 Support of low rank matrices . -

This Booklet
Applied Mathematics Department of Mathematical Sciences Research Projects 2018-2019 2 Forecasting solar and wind power outputs using deep learning approaches Research Team: Dr Bubacarr Bah In collaboration with University of Pretoria Highlights Identification of suitable locations of wind and solar farms in South Africa. Prediction of wind (solar) power from historical wind (solar) data, and other environmental factors, of South Africa using recurrent neural networks. Kalle Pihlajasaari / Wikimedia Commons / CC-BY-SA-3.0 Figure. Wind turbines at Darling, Western Cape. Applications Informed decision making by power producer in terms of integra- tion of renewable energies into existing electricity grids. Forecasting of energy prices. 3 Machine learning outcome prediction for coma patients Research Team: Dr Bubacarr Bah In collaboration with University of Cape Town Highlights Outcome prediction with Serial Neuron-Specific Enolase (SNSE) measurements from coma patients. The use of k-nearest neighbors (k-NN) for imputation of missing data shows promising results. Outcome prediction with EEG signals from coma patients. Using machine learning to determine the prognostic power of SNSE and EEG Figure. EEG and the determination of level of consciousness in coma patients. Applications Potential application in medical and health systems to improve diag- nosis (and treatment) of coma. 4 Data-driven river flow routing using deep learning Research Team: Dr Willie Brink and Prof Francois Smit MSc Student: Jaco Briers Highlights Predicting flow along the lower Orange River in South Africa, for improved release scheduling at the Vanderkloof Dam, using recur- rent neural networks as well as feedforward convolutional neural networks trained on historical time series records. -

Tuesday July 9, 1.45-8.00 Pm
Tuesday July 9, 1.45-8.00 pm 1.45-2.45 Combinatorial compressed sensing with expanders Bubacarr Bah Wegener Chair: Ana Gilbert Deep learning (invited session) Chair: Misha Belkin & Mahdi Soltanolkotabi 2:55-3:20 Reconciling modern machine learning practice and the classical bias-variance trade-off Mikhail Belkin, Daniel Hsu, Siyuan Ma & Soumik Mandal 3.20-3.45 Overparameterized Nonlinear Optimization with Applications to Neural Nets Samet Oymak 3.45-4.10 General Bounds for 1-Layer ReLU approximation Bolton R. Bailey & Matus Telgarsky 4.10-4.20 Short Break A9 Amphi 1 4.20-4.45 Generalization in deep nets: an empirical perspective Tom Goldstein 4.45-5.10 Neuron birth-death dynamics accelerates gradient descent and converges asymptotically Joan Bruna 5.45-8.00 Poster simposio Domaine du Haut-Carr´e Tuesday July 9, 1.45-8.00 pm 1.45-2.45 Combinatorial compressed sensing with expanders Bubacarr Bah Wegener Chair: Anna Gilbert Frame Theory Chair: Ole Christensen 2.55-3.20 Banach frames and atomic decompositions in the space of bounded operators on Hilbert spaces Peter Balazs 3.20-3.45 Frames by Iterations in Shift-invariant Spaces Alejandra Aguilera, Carlos Cabrelli, Diana Carbajal & Victoria Paternostro 3.45-4.10 Frame representations via suborbits of bounded operators Ole Christensen & Marzieh Hasannasabjaldehbakhani 4.10-4.20 Short Break 4.20-4.45 Sum-of-Squares Optimization and the Sparsity Structure A29 Amphi 2 of Equiangular Tight Frames Dmitriy Kunisky & Afonso Bandeira 4.45-5.10 Frame Potentials and Orthogonal Vectors Josiah Park 5.10-5.35 -
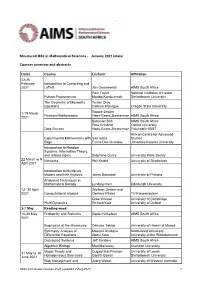
Structured Msc in Mathematical Sciences - January 2021 Intake
` Structured MSc in Mathematical Sciences - January 2021 intake Courses overview and abstracts Dates Course Lecturer Affiliation 22-26 February Introduction to Computing and 2021 LaTeX Jan Groenewald AIMS South Africa Paul Taylor National Institutes of Health Python Programming Martha Kamkuemah Stellenbosch University The Geometry of Maxwell’s Tevian Dray Equations Corinne Manogue Oregon State University 1-19 March Ronnie Becker Financial Mathematics Hans-Georg Zimmerman AIMS South Africa 2021 Bubacarr Bah AIMS South Africa Pete Grindrod Oxford University Data Science Hans-Georg Zimmerman Fraunhofer IBMT African Centre for Advanced Experimental Mathematics with Yae Gaba Studies Sage Evans Doe Ocansey Johannes Keppler University Introduction to Random Systems, Information Theory, and related topics Stéphane Ouvry Université Paris Saclay 22 March to 9 Networks Phil Knight University of Strathclyde April 2021 Introduction to Multiscale Models and their Analysis Jacek Banasiak University of Pretoria Analytical Techniques in Mathematical Biology Lyndsay Kerr Edinburgh University 12- 30 April Wolfram Decker and 2021 Computational Algebra Gerhard Pfister TU Kaiserslautern Grae Worster University of Cambridge Fluid Dynamics Richard Katz University of Oxford 3-7 May Reading week 10-28 May Probability and Statistics Daniel Nickelsen AIMS South Africa 2021 Biophysics at the Microscale Daisuke Takagi University of Hawaii at Manoa Symmetry Analysis of Masood Khalique North-West University Differential Equations Abdul Kara University of the Witwatersrand -

Visualization and Analysis of Single-Cell Data Using Diffusion Maps
Visualization and Analysis of Single-Cell Data using Diffusion Maps Lisa Mariah Beer Born 23rd December 1994 in Suhl, Germany 24th September 2018 Master’s Thesis Mathematics Advisor: Prof. Dr. Jochen Garcke Second Advisor: Prof. Dr. Martin Rumpf Institute for Numerical Simulation Mathematisch-Naturwissenschaftliche Fakultat¨ der Rheinischen Friedrich-Wilhelms-Universitat¨ Bonn Contents Symbols and Abbreviations 1 1 Introduction 3 2 Single-Cell Data 5 3 Data Visualization 9 3.1 Dimensionality Reduction . .9 3.2 Diffusion Maps . 10 3.2.1 Bandwidth Selection . 14 3.2.2 Gaussian Kernel Selection . 16 3.3 Experiments . 17 4 Group Detection 25 4.1 Spectral Clustering . 25 4.2 Diffusion Pseudotime Analysis . 27 4.2.1 Direct Extension . 30 4.3 Experiments . 31 5 Consideration of Censored Data 43 5.1 Gaussian Kernel Estimation . 43 5.2 Euclidean Distance Estimation . 45 5.3 Experiments . 46 6 Conclusion and Outlook 51 A Appendix 53 References 63 i ii Symbols and Abbreviations Symbols N Set of natural numbers R Set of real numbers m d m is much smaller than d k · k2 Euclidean norm G = (V, E) A graph consisting of a set of vertices V and (weighted) edges E 1 A vector with ones diag(v) A diagonal matrix with v being the diagonal ρ(A) The spectral radius of a matrix A O(g) Big O notation χI The indicator function of a set I erfc The Gaussian complementary error function E[X] The expectation of a random variable X V ar[X] The variance of a random variable X |X| The cardinality of a set X X A given data set M The manifold, the data is lying on n The number -

10 Diffusion Maps
10 Diffusion Maps - a Probabilistic Interpretation for Spectral Embedding and Clustering Algorithms Boaz Nadler1, Stephane Lafon2,3, Ronald Coifman3,and Ioannis G. Kevrekidis4 1 Department of Computer Science and Applied Mathematics, Weizmann Institute of Science, Rehovot, 76100, Israel, [email protected] 2 Google, Inc. 3 Department of Mathematics, Yale University, New Haven, CT, 06520-8283, USA, [email protected] 4 Department of Chemical Engineering and Program in Applied and Computational Mathematics, Princeton University, Princeton, NJ 08544, USA, [email protected] Summary. Spectral embedding and spectral clustering are common methods for non-linear dimensionality reduction and clustering of complex high dimensional datasets. In this paper we provide a diffusion based probabilistic analysis of al- gorithms that use the normalized graph Laplacian. Given the pairwise adjacency matrix of all points in a dataset, we define a random walk on the graph of points and a diffusion distance between any two points. We show that the diffusion distance is equal to the Euclidean distance in the embedded space with all eigenvectors of the normalized graph Laplacian. This identity shows that characteristic relaxation times and processes of the random walk on the graph are the key concept that gov- erns the properties of these spectral clustering and spectral embedding algorithms. Specifically, for spectral clustering to succeed, a necessary condition is that the mean exit times from each cluster need to be significantly larger than the largest (slowest) of all relaxation times inside all of the individual clusters. For complex, multiscale data, this condition may not hold and multiscale methods need to be developed to handle such situations. -

Diffusion Maps and Transfer Subspace Learning
Wright State University CORE Scholar Browse all Theses and Dissertations Theses and Dissertations 2017 Diffusion Maps and Transfer Subspace Learning Olga L. Mendoza-Schrock Wright State University Follow this and additional works at: https://corescholar.libraries.wright.edu/etd_all Part of the Computer Engineering Commons, and the Computer Sciences Commons Repository Citation Mendoza-Schrock, Olga L., "Diffusion Maps and Transfer Subspace Learning" (2017). Browse all Theses and Dissertations. 1845. https://corescholar.libraries.wright.edu/etd_all/1845 This Dissertation is brought to you for free and open access by the Theses and Dissertations at CORE Scholar. It has been accepted for inclusion in Browse all Theses and Dissertations by an authorized administrator of CORE Scholar. For more information, please contact [email protected]. DIFFUSION MAPS AND TRANSFER SUBSPACE LEARNING A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy by OLGA L. MENDOZA-SCHROCK B.S., University of Puget Sound, 1998 M.S., University of Kentucky, 2004 M.S., Wright State University, 2013 2017 Wright State University WRIGHT STATE UNIVERSITY GRADUATE SCHOOL July 25, 2017 I HEREBY RECOMMEND THAT THE DISSERTATION PREPARED UNDER MY SUPERVISION BY Olga L. Mendoza-Schrock ENTITLED Diffusion Maps and Transfer Subspace Learning BE ACCEPTED IN PARTIAL FULFILL- MENT OF THE REQUIREMENTS FOR THE DEGREE OF Doctor of Philosophy Mateen M. Rizki, Ph.D. Dissertation Director Michael L. Raymer, Ph.D. Director, Ph.D. in Computer Science and Engineering Program Robert E. W. Fyffe, Ph.D. Vice President for Research and Dean of the Graduate School Committee on Final Examination John C. -

Diffusion Maps and Transfer Subspace Learning
DIFFUSION MAPS AND TRANSFER SUBSPACE LEARNING A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy by OLGA L. MENDOZA-SCHROCK B.S., University of Puget Sound, 1998 M.S., University of Kentucky, 2004 M.S., Wright State University, 2013 2017 Wright State University WRIGHT STATE UNIVERSITY GRADUATE SCHOOL July 25, 2017 I HEREBY RECOMMEND THAT THE DISSERTATION PREPARED UNDER MY SUPERVISION BY Olga L. Mendoza-Schrock ENTITLED Diffusion Maps and Transfer Subspace Learning BE ACCEPTED IN PARTIAL FULFILL- MENT OF THE REQUIREMENTS FOR THE DEGREE OF Doctor of Philosophy Mateen M. Rizki, Ph.D. Dissertation Director Michael L. Raymer, Ph.D. Director, Ph.D. in Computer Science and Engineering Program Robert E. W. Fyffe, Ph.D. Vice President for Research and Dean of the Graduate School Committee on Final Examination John C. Gallagher, Ph.D. Frederick D. Garber, Ph.D. Michael L. Raymer, Ph.D. Vincent J. Velten, Ph.D. ABSTRACT Mendoza-Schrock, Olga L., Ph.D., Computer Science and Engineering Ph.D. Program, De- partment of Computer Science and Engineering, Wright State University, 2017. Diffusion Maps and Transfer Subspace Learning. Transfer Subspace Learning has recently gained popularity for its ability to perform cross-dataset and cross-domain object recognition. The ability to leverage existing data without the need for additional data collections is attractive for Aided Target Recognition applications. For Aided Target Recognition (or object assessment) applications, Trans- fer Subspace Learning is particularly useful, as it enables the incorporation of sparse and dynamically collected data into existing systems that utilize large databases. -
![Arxiv:1506.07840V1 [Stat.ML] 25 Jun 2015 Are Nonlinear, Which Is Essential As Real-World Data Typically Does Not Lie on a Hyperplane](https://docslib.b-cdn.net/cover/7375/arxiv-1506-07840v1-stat-ml-25-jun-2015-are-nonlinear-which-is-essential-as-real-world-data-typically-does-not-lie-on-a-hyperplane-5447375.webp)
Arxiv:1506.07840V1 [Stat.ML] 25 Jun 2015 Are Nonlinear, Which Is Essential As Real-World Data Typically Does Not Lie on a Hyperplane
Diffusion Nets Gal Mishne, Uri Shaham, Alexander Cloninger and Israel Cohen∗ June 26, 2015 Abstract Non-linear manifold learning enables high-dimensional data analysis, but requires out- of-sample-extension methods to process new data points. In this paper, we propose a manifold learning algorithm based on deep learning to create an encoder, which maps a high-dimensional dataset and its low-dimensional embedding, and a decoder, which takes the embedded data back to the high-dimensional space. Stacking the encoder and decoder together constructs an autoencoder, which we term a diffusion net, that performs out-of- sample-extension as well as outlier detection. We introduce new neural net constraints for the encoder, which preserves the local geometry of the points, and we prove rates of con- vergence for the encoder. Also, our approach is efficient in both computational complexity and memory requirements, as opposed to previous methods that require storage of all train- ing points in both the high-dimensional and the low-dimensional spaces to calculate the out-of-sample-extension and the pre-image. 1 Introduction Real world data is often high dimensional, yet concentrates near a lower dimensional mani- fold, embedded in the high dimensional ambient space. In many applications, finding a low- dimensional representation of the data is necessary to efficiently handle it and the representation usually reveals meaningful structures within the data. In recent years, different manifold learn- ing methods have been developed for high dimensional data analysis, which are based on the geometry of the data, i.e. preserving distances within local neighborhoods of the data. -

Variational Diffusion Autoencoders with Random
Under review as a conference paper at ICLR 2020 VARIATIONAL DIFFUSION AUTOENCODERS WITH RANDOM WALK SAMPLING Anonymous authors Paper under double-blind review ABSTRACT Variational inference (VI) methods and especially variational autoencoders (VAEs) specify scalable generative models that enjoy an intuitive connection to manifold learning — with many default priors the posterior/likelihood pair q(zjx)/p(xjz) can be viewed as an approximate homeomorphism (and its inverse) between the data manifold and a latent Euclidean space. However, these approx- imations are well-documented to become degenerate in training. Unless the sub- jective prior is carefully chosen, the topologies of the prior and data distributions often will not match. Conversely, diffusion maps (DM) automatically infer the data topology and enjoy a rigorous connection to manifold learning, but do not scale easily or provide the inverse homeomorphism. In this paper, we propose a) a principled measure for recognizing the mismatch between data and latent dis- tributions and b) a method that combines the advantages of variational inference and diffusion maps to learn a homeomorphic generative model. The measure, the locally bi-Lipschitz property, is a sufficient condition for a homeomorphism and easy to compute and interpret. The method, the variational diffusion autoen- coder (VDAE), is a novel generative algorithm that first infers the topology of the data distribution, then models a diffusion random walk over the data. To achieve efficient computation in VDAEs, we use stochastic versions of both variational inference and manifold learning optimization. We prove approximation theoretic results for the dimension dependence of VDAEs, and that locally isotropic sam- pling in the latent space results in a random walk over the reconstructed manifold. -

Bubacarr Bah CV
Curriculum Vitae of Bubacarr Bah Address African Institute for Mathematical Sciences (AIMS) South Africa 6 Melrose Road Muizenberg 7945 South Africa Recent Employment History Oct 2016 { present German Research Chair (Senior Researcher) in Mathematics of Data Science, AIMS South Africa, & Senior Lecturer (Asst. Prof.), Stellenbosch University Sep 2014 { Sep 2016 Postdoctoral Fellow, Mathematics Department, & Institute for Computational Engineering & Sciences, University of Texas in Austin Sep 2012 { Aug 2014 Research Scientist (postdoc), Laboratory for Information and Inference Systems, Ecole Polytechnique Federale de Lausanne (EPFL) Education Sep 2008 { Aug 2012 PhD in Applied and Computational Mathematics, University of Edinburgh, UK Sep 2007 { Sep 2008 MSc in Mathematical Modelling & Scientific Computing, University of Oxford, UK Jan 2000 { Aug 2004 BSc in Mathematics and Physics, (summa cum laude), University of The Gambia Honours 2020 Alexander von Humboldt Award of German Research Chair Mathematics at AIMS South Africa 2016 Alexander von Humboldt Award of German Research Chair Mathematics at AIMS South Africa 2015 NSF-funded SIAM ICIAM15 Travel Award 2013 Travel Grant for Matheon Workshop 2013 on Compressed Sensing & its Applications 2011 SIAM Certificate of Recognition 2010 SIAM Best Student Paper Prize 2008 Edinburgh University Principal's Scholarship for PhD studies 2007 Commonwealth Scholarship for MSc studies 2005 Overall Best Student (valedictorian) Award 2005 Best Student of the Faculty of Science & Agriculture Award 2005 Vice Chancellor's Award Journal Publications 1. Improved restricted isometry constant bounds for Gaussian matrices; SIAM Journal on Matrix Analysis, Vol. 31(5) (2010) 2882{2898 (with J. Tanner). 2. Bounds of restricted isometry constants in extreme asymptotics: formulae for Gaussian matrices; Linear Algebra and its Applications, Vol.