Testing the Reliability of Standard and Complementary DNA Barcodes for the Monocot Subfamily Alooideae from South Africa Barnabas H
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
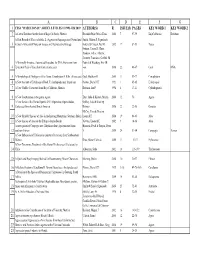
Haseltonia Articles and Authors.Xlsx
ABCDEFG 1 CSSA "HASELTONIA" ARTICLE TITLES #1 1993–#26 2019 AUTHOR(S) R ISSUE(S) PAGES KEY WORD 1 KEY WORD 2 2 A Cactus Database for the State of Baja California, Mexico Resendiz Ruiz, María Elena 2000 7 97-99 BajaCalifornia Database A First Record of Yucca aloifolia L. (Agavaceae/Asparagaceae) Naturalized Smith, Gideon F, Figueiredo, 3 in South Africa with Notes on its uses and Reproductive Biology Estrela & Crouch, Neil R 2012 17 87-93 Yucca Fotinos, Tonya D, Clase, Teodoro, Veloz, Alberto, Jimenez, Francisco, Griffith, M A Minimally Invasive, Automated Procedure for DNA Extraction from Patrick & Wettberg, Eric JB 4 Epidermal Peels of Succulent Cacti (Cactaceae) von 2016 22 46-47 Cacti DNA 5 A Morphological Phylogeny of the Genus Conophytum N.E.Br. (Aizoaceae) Opel, Matthew R 2005 11 53-77 Conophytum 6 A New Account of Echidnopsis Hook. F. (Asclepiadaceae: Stapeliae) Plowes, Darrel CH 1993 1 65-85 Echidnopsis 7 A New Cholla (Cactaceae) from Baja California, Mexico Rebman, Jon P 1998 6 17-21 Cylindropuntia 8 A New Combination in the genus Agave Etter, Julia & Kristen, Martin 2006 12 70 Agave A New Series of the Genus Opuntia Mill. (Opuntieae, Opuntioideae, Oakley, Luis & Kiesling, 9 Cactaceae) from Austral South America Roberto 2016 22 22-30 Opuntia McCoy, Tom & Newton, 10 A New Shrubby Species of Aloe in the Imatong Mountains, Southern Sudan Leonard E 2014 19 64-65 Aloe 11 A New Species of Aloe on the Ethiopia-Sudan Border Newton, Leonard E 2002 9 14-16 Aloe A new species of Ceropegia sect. -

Checklist of the Vascular Alien Flora of Catalonia (Northeastern Iberian Peninsula, Spain) Pere Aymerich1 & Llorenç Sáez2,3
BOTANICAL CHECKLISTS Mediterranean Botany ISSNe 2603-9109 https://dx.doi.org/10.5209/mbot.63608 Checklist of the vascular alien flora of Catalonia (northeastern Iberian Peninsula, Spain) Pere Aymerich1 & Llorenç Sáez2,3 Received: 7 March 2019 / Accepted: 28 June 2019 / Published online: 7 November 2019 Abstract. This is an inventory of the vascular alien flora of Catalonia (northeastern Iberian Peninsula, Spain) updated to 2018, representing 1068 alien taxa in total. 554 (52.0%) out of them are casual and 514 (48.0%) are established. 87 taxa (8.1% of the total number and 16.8 % of those established) show an invasive behaviour. The geographic zone with more alien plants is the most anthropogenic maritime area. However, the differences among regions decrease when the degree of naturalization of taxa increases and the number of invaders is very similar in all sectors. Only 26.2% of the taxa are more or less abundant, while the rest are rare or they have vanished. The alien flora is represented by 115 families, 87 out of them include naturalised species. The most diverse genera are Opuntia (20 taxa), Amaranthus (18 taxa) and Solanum (15 taxa). Most of the alien plants have been introduced since the beginning of the twentieth century (70.7%), with a strong increase since 1970 (50.3% of the total number). Almost two thirds of alien taxa have their origin in Euro-Mediterranean area and America, while 24.6% come from other geographical areas. The taxa originated in cultivation represent 9.5%, whereas spontaneous hybrids only 1.2%. From the temporal point of view, the rate of Euro-Mediterranean taxa shows a progressive reduction parallel to an increase of those of other origins, which have reached 73.2% of introductions during the last 50 years. -

Prickly News South Coast Cactus & Succulent Society Newsletter | April 2021
PRICKLY NEWS SOUTH COAST CACTUS & SUCCULENT SOCIETY NEWSLETTER | APRIL 2021 Karen ZOOM PRESENTATION SHARE YOUR GARDEN OR YOUR FAVORITE PLANT Ostler Sunday, April 14 @ 1:30 pm “Staging” Happy Spring! The cactus potting season is upon us. It is time to repot and spruce up Email me with photos of your garden and/or plants those plants that no longer fit their that we can publish as a way of staying connected. containers. [email protected] Our upcoming meeting will give you some special tips on potting and staging your plants. I always enjoy Karen Ostler’s approach to staging To learn more visit southcoastcss.org and I hope you will also. Like us on our facebook page CALL FOR PHOTOS Please E-mail photos of your favorite cacti and/or succulents to me at Follow us on Instagram, _sccss_ [email protected] by April 9th so I may present them at the upcoming Zoom meeting. If you have a plant in the ground or want to share IN THIS ISSUE your garden, those photos are also welcome. Please join in on Sunday, April 11th at 1 PM for PRESENTER OF THE MONTH 2 social half hour and 1:30 PM for the presentation. WHAT’S IN YOUR GARDEN? 3 TIP OF THE MONTH 6 Stay well and keep in touch! MINI-SHOW PLANT CALENDAR 8 MARIA CAPALDO MINISHOW SUCCULENT WINNERS 9 MINISHOW CACTUS WINNERS 13 LATIN LOOKUP 16 MINI SHOW MARCH PLANTS 18 MINI SHOW STANDINGS 24 CANCELLED EVENTS 25 BOARD OF DIRECTORS 25 1 APRIL SCCSS Presenter of the month “Staging” BY KAREN OSTLER Karen has been growing cactus since she got her first ‘real’ cactus, a Gymnocalycium friedrichii, now G. -

Succulents-Plant-List-2021.Pdf
Rutgers Gardens Spring Plant Sale 2021 ‐ SUCCULENTS (all plants available from May 1) Scientific name Cultivar name, notes Common name Adromischus cristatus crinkle‐leaf plant, key lime pie Aeonium percarneum kiwi aeonium Agave americana century plant Agave americana Marginata century plant Agave montana Agave schidigera (Agave filifera var. schidigera) Aloe Delta Lights Aloe arborescens Octopus Aloe Bulbine frutescens Hallmark Coprosma Evening Glow mirror plant Crassula Tom Thumb Crassula Small Red Carpet Crassula falcata propeller plant Crassula ovata Gollum jade tree Crassula ovata Hummel's Sunset golden jade tree Crassula pellucida Variegata calico kitten crassula Crassula perforata string of buttons Cremnosedum Little Gem Delosperma echinatum pickle plant Disocactus anguliger Epiphyllum anguliger fishbone cactus, zig zag cactus Echeveria Pearl Von Nurmberg Echeveria Elegans hens and chicks Echeveria Woolly Rose hens and chicks Echeveria gibbiflora Echeveria nodulosa Echeveria runyonii Topsy Turvy Echeveria setosa Euphorbia Sticks on Fire red pencil tree, fire sticks Euphorbia lactea f. cristata coral cactus Euphorbia mammillaris indian corn cob Euphorbia milii dwarf crown of thorns Euphorbia milii crown of thorns Faucaria tuberculosa tiger jaws Gasteria Little Warty Graptopetalum paraguayense mother‐of‐pearl‐plant, ghost plant Graptosedum Vera Higgins Graptosedum Darley Sunshine Haworthiopsis attenuata var. Big Band zebra plant Haworthiopsis tessellata (Haworthia t.) Haworthiopsis venosa (Haworthia v.) Kalanchoe Silver Spoons Kalanchoe -

Koenabib Mine Near Aggeneys, Northern Cape Province
KOENABIB MINE NEAR AGGENEYS, NORTHERN CAPE PROVINCE BOTANICAL STUDY AND ASSESSMENT Version: 1.0 Date: 30th January 2020 Authors: Gerhard Botha & Dr. Jan -Hendrik Keet PROPOSED MINING OF SILLIMANITE, AGGREGATE AND GRAVEL ON THE FARM KOENABIB 43 NORTH OF AGGENEYS, NORTHERN CAPE PROVINCE Report Title: Botanical Study and Assessment Authors: Mr. Gerhard Botha & Dr. Jan-Hendrik Keet Project Name: Proposed Mining of Sillimanite, Aggregate and Gravel on the Farm Koenabib 43, North of Aggeneys, Northern Cape Province Status of report: Version 1.0 Date: 30th January 2020 Prepared for: Greenmined Environmental Postnet Suite 62, Private Bag X15 Somerset West 7129 Cell: 082 734 5113 Email: [email protected] Prepared by Nkurenkuru Ecology and Biodiversity 3 Jock Meiring Street Park West Bloemfontein 9301 Cell: 083 412 1705 Email: gabotha11@gmail com Suggested report citation Nkurenkuru Ecology and Biodiversity, 2019. Mining Permit, Final Basic Assessment & Environmental Management Plan for the proposed mining of Sillimanite, Aggregate and Stone Gravel on the Farm Koenabib 43, Northern Cape Province. Botanical Study and Assessment Report. Unpublished report prepared by Nkurenkuru Ecology and Biodiversity for GreenMined Environmental. Version 1.0, 30 January 2020. Proposed koenabib sillimanite mine, NORTHERN CAPE PROVINCE January 2020 botanical STUDY AND ASSESSMENT I. DECLARATION OF CONSULTANTS INDEPENDENCE » act/ed as the independent specialist in this application; » regard the information contained in this report as it relates to my specialist -

Opuntia Farm in Syria by Fouad Shalghin
Vol. 57, No. 3 May-June 2020 Opuntia Farm in Syria www.hcsstex.org by Fouad Shalghin 1 Vol. 57, No. 3 May-June 2020 From the editor Karla Halpaap-Wood I want to thank everybody who contributed to this issue of the KK, especially Chaden Yafi for her interesting article. My big thanks goes also to Irwin Lightstone from NTCSS for introducing me to Zoom meetings and being very helpful with practical advice. MEMBERSHIP KATHY FEWOX & JULY OLSON Due to coronavirus social distancing, both the March and April meetings at the Metropolitan Multi-Service Center had to be cancelled. So was everything fun we had planned for this part of the year. Big Bend field trip, open gardens, Spring Sale, potting party — all gone with the pandemic. However, as disappointed as we all were, it had to be done. Nobody wants to become ill, or cause someone else to get sick. On the bright side, the April membership meeting was held via Zoom. Twelve members took part. We did not have an official program but three plants of the month were presented, one from March when the meeting was cancelled and the two plants from April. Presentations were very nice and pictures and plants could be seen clearly. Sadly, two members of the club recently lost loved ones. David Van Langen’s father, Burk, passed away on April 28, only a few months after David’s mother’s death. After an eight-year-long illness, starting with cancer and most recently vascular dementia, Liliana Cracraft’s mother Maria Angelica Treviño (Keka) passed away on March 2. -

CREW Newsletter – 2021
Volume 17 • July 2021 Editorial 2020 By Suvarna Parbhoo-Mohan (CREW Programme manager) and Domitilla Raimondo (SANBI Threatened Species Programme manager) May there be peace in the heavenly virtual platforms that have marched, uninvited, into region and the atmosphere; may peace our homes and kept us connected with each other reign on the earth; let there be coolness and our network of volunteers. in the water; may the medicinal herbs be healing; the plants be peace-giving; may The Custodians of Rare and Endangered there be harmony in the celestial objects Wildflowers (CREW), is a programme that and perfection in eternal knowledge; may involves volunteers from the public in the everything in the universe be peaceful; let monitoring and conservation of South peace pervade everywhere. May peace abide Africa’s threatened plants. CREW aims to in me. May there be peace, peace, peace! capacitate a network of volunteers from a range of socio-economic backgrounds – Hymn of peace adopted to monitor and conserve South Africa’s from Yajur Veda 36:17 threatened plant species. The programme links volunteers with their local conservation e are all aware that our lives changed from the Wend of March 2020 with a range of emotions, agencies and particularly with local land from being anxious of not knowing what to expect, stewardship initiatives to ensure the to being distressed upon hearing about friends and conservation of key sites for threatened plant family being ill, and sometimes their passing. De- species. Funded jointly by the Botanical spite the incredible hardships, we have somehow Society of South Africa (BotSoc), the Mapula adapted to the so-called new normal of living during Trust and the South African National a pandemic and are grateful for the commitment of the CREW network to continue conserving and pro- Biodiversity Institute (SANBI), CREW is an tecting our plant taxa of conservation concern. -

2020 MSU Horticulture Gardens Houseplant and Succulent Sale - Tentative Inventory
2020 MSU Horticulture Gardens Houseplant and Succulent Sale - Tentative Inventory Scroll Down for Cacti and Foliage Plants All Prices- To Be Determined Item Code Quantity Pot Size Genus species Common Name Family Light Water Category SUCCULENTS SUCCULENTS SUCCULENTS SUCCULENTS SUCCULENTS SUCCULENTS SUCCULENTS SUCCULENTS SUCCULENTS SUCCULENTS SUCCULENTS SUCCULENTS ADRO001 4" to 4.5" Adromischus cristatus 'Key Lime Pie' Crinkle-leaf Plant Crassulaceae High Low Succulent AEON001 15 4" Aeonium arborescens ' Tip Top' Dwarf Tree Aeonium Crassulaceae High Low Succulent AEON002 10 4" Aeonium atropurpureum Purple Aeonium Crassulaceae High Low Succulent AEON003 4" Aeonium castello-paivae variegata 'Suncup' Suncup Aeonium Crassulaceae High Low Succulent AEON004 30 4" Aeonium haworthii 'Kiwi' Haworth's Aeonium Crassulaceae High Low Succulent AEON005 15 4" Aeonium hybrid 'Stripe' Stripe Aeonium Crassulaceae High Low Succulent AGAV001 3 4" Agave geminiflora Twin flowered Agave Agavoideae High Low Succulent AGAV002 43 4.5" Agave gypsophyla 'Ivory Curls' Ivory Curls Century Plant Agavoideae High Low Succulent AGAV003 39 3.5" to 4" Agave victoriae-reginae 'Porcupine' Queen Victoria Agave Agavoideae High Low Succulent AGAV004 64 4.5" Agave xylonacantha 'Frostbite' Frostbite Century Plant Agavoideae High Low Succulent ALBU001 14 6" Albuca bracteata Pregnant Onion Asparagaceae High Low Succulent ALBU002 36 4" Albuca bracteata Pregnant Onion Asparagaceae High Low Succulent ALOE001 33 4" Aloe aristata Lace Aloe Asphodelaceae High Low Succulent ALOE002 26 4" Aloe ciliaris Climbing Aloe Asphodelaceae High Low Succulent ALOE003 20 4" Aloe hybrid 'Minnie Belle' Hybrid Aloe Asphodelaceae High Low Succulent ALOE004 37 4" Aloe hybrid 'Pink Blush' Pink Blush Aloe Asphodelaceae High Low Succulent ALOE005 3 6" Aloe hybrid 'T. -

Alsterworthia International. Volume 18, Issue 3 Contents
Haworthia retusa at Kruisrivierm, SA. Photograph Bruce Bayer Contents. Alsterworthia Internaonal ‐ renewal details etc…………………………………………………………………………………………………………….2 Nomina Nova recorded for the Asphodelaceae for 2015 and 2016 in the Repertorium Plantarum Succulentarum. …………………………………………………………………………………………………….. …………. 3‐10 Japanese Haworthia Society, Haworthia Study, Editor Dr Hayashi………………………..………………………………………..…………….10 Internaonal Succulent Introducons 2018. Plant Introducons of the Hunngton Botanical Gardens…………………..11‐25 Some thoughts on our plants………………………………………………………………………………………………………………………………...…26‐27 Variaon in Haworthia retusa at Kruisrivier, South Africa. ……………………………………………………………… ..…Front page.& back Vol. 18 Issue 3, November 2018 ISSN1474‐4635 Alsterworthia International. Volume 18, Issue 3 Alsterworthia Internaonal Editor: Harry Mays BSc, BA (Honours), Member IOS. Woodsleigh, Moss Lane, St. Michaels on Wyre, Preston, PR3 0TY, U.K. E‐Mail: [email protected]. Renewal subscripon for 2019. UK = £14.00. Rest of the world £19.00, surface postage included (for airmail add £4.00). Payments in Brish pounds only please: 1. by PayPal to: [email protected] Add 4% to compensate Alsterworthia for deducon made by PayPal. 2. by Bank transfers, as appropriate, to: Santander, account Alsterworthia Internaonal. Account number 75923083. Sort Code 090155. BIC: ABBYGB21XXX. IBAN: GB8OABBY09015575923083. If paying by bank transfer please ensure you pay ALL bank charges (i.e. senders and receivers). 3. by cheques, bank dras payable to “Alsterworthia Internaonal” ‐ please send to: H. Mays, Woodsleigh, Moss Lane, St Michaels on Wyre, Preston, PR3 0TY, UK. 4. Members in Belgium may pay subscripon in local currency to Frank Thys, Antwerpsedreef 30, B‐2980, Zoersel, Belgium. E‐mail: frank‐[email protected] in Australia to Paul Forster, P.O. Box 2171, Ashgrove, West Queensland 4060, Australia. -

Prickly News South Coast Cactus & Succulent Society Newsletter | February 2020
PRICKLY NEWS SOUTH COAST CACTUS & SUCCULENT SOCIETY NEWSLETTER | FEBRUARY 2020 NEXT MEETING REFRESHMENTS OUR MISSION Sunday, Feb 9, 2020 Thank you... We Aim To: Sean Dean Gildenhuys to those who brought Promote education and refreshments in January. cooperation among hobby- Haworthiopsis ists and others interested Jim Gardner in the study, culture, and Maria Capaldo propagation of succulent plants, including cacti. Marie Bowers It is time to start preparing for this year’s Support the activities of Show and Sale which will be April 4-5, 2020, Judy Unrine organizations whose goals the first weekend in April. Please put this on Clif Wong include the conservation your calendar. of cacti and succulents in Please bring something their native habitats. At the February meeting, we will be recruiting yummy to share for our members for various positions. For the Show, Learn facts, make you may decide to have a display table, of either February meeting... friends, have fun! artistic or specimen plants. Usually members bring in their best plants or a theme or one species to place on their table. See Carol Knight, Show and Sale Chair, to arrange this. Also, check your To learn more visit southcoastcss.org plant collection to see if you have some plants you can get ready for the Society Display Tables. You will receive a Mini-show point for each plant up to 4 for display plants or 4 points for a display table. Like us on our facebook page If you’re new to the society, we will explain in some detail these activities at the meeting. -

Special Issue No 11 (New Combinations)
Some new combinaons in Haworthia. Haworthiopsis. Tulista. by Ingo Breuer. Graf‐von‐Galen‐Str. 105 52525 Heinsberg Germany. E‐mail: IBreuer@t‐online.de Web site: www.eden‐plants.com Haworthia badia var. joleneae. IB1892 Contents. Some new combinations in Haworthia, Haworthiopsis and Tulista. Ingo Breuer, Germany ................................................... 3-4 New Names in Haworthia, Haworthiopsis and Tulista. Ingo Breuer ....................................................................................... 5-7 Accepted Names in Haworthia, Haworthiopsis and Tulista. Ingo Breuer .............................................................................. 7-11 References................................................................................................................................................................................. 12 Genus Haworthia. Photographs of each species in alphabetical order. ............................................................................... 13-24 Genus Haworthiopsis. Photographs of each species in alphabetical order. ......................................................................... 24-26 Genus Tulista. Photographs of each species in alphabetical order. ...................................................................................... 26-27 Enlargements of selected photographs ....................................................................................................... Front and back covers 25 June 2016 Alsterworthia Internaonal Special Issue No. 11 ISSN: -

Haworthiopsis Limifolia (Asphodelaceae): Medicinal Uses, Phytochemistry and Biological Activities
Alfred Maroyi /J. Pharm. Sci. & Res. Vol. 11(9), 2019, 3366-3371 Haworthiopsis limifolia (Asphodelaceae): medicinal uses, phytochemistry and biological activities Alfred Maroyi Medicinal Plants and Economic Development (MPED) Research Centre, Department of Botany, University of Fort Hare, Private Bag X1314, Alice 5700, South Africa Abstract Haworthiopsis limifolia is a succulent herbaceous perennial plant widely used as herbal medicine in southern Africa. This study reviewed medicinal uses, phytochemistry and pharmacological properties of H. limifolia. Relevant information on the uses, phytochemistry and pharmacological properties of H. limifolia was collected from electronic scientific databases such as ScienceDirect, SciFinder, PubMed, Google Scholar, Medline, and SCOPUS. Pre-electronic literature search of conference papers, scientific articles, books, book chapters, dissertations and theses was carried out at the University library. Literature search revealed that H. limifolia is used as a protective charm against enemies, evil spirits, lightning, snakes, storms, and bring good fortune or luck. The leaves, roots and whole plant parts of H. limifolia are used as herbal medicines for fertility problems, sores, purifying blood, cough, skin rashes, sun burns, burns and gastro-intestinal problems. Phytochemical compounds identified from the species include alkaloids, anthraquinones, carbohydrates, fatty acids, flavonoids, glycosides, lectins, phenolics, saponins, sterols and tannins. Pharmacological studies revealed that H. limifolia extracts