The Roles of Host Noncoding Rnas in Mycobacterium Tuberculosis Infection
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Autophagy: from Basic Science to Clinical Application
nature publishing group REVIEW See COMMENTARY page XX Autophagy: from basic science to clinical application J Va n L i m b e r g e n 1 , 2 , 3 , C S t e v e n s 4 , E R N i m m o 1 , D C W i l s o n 2 , 3 a n d J S a t s a n g i 1 Autophagy is a cellular pathway involved in protein and organelle degradation, which is likely to represent an innate adaptation to starvation. In times of nutrient deficiency, the cell can self-digest and recycle some nonessential components through nonselective autophagy, thus sustaining minimal growth requirements until a food source becomes available. Over recent years, autophagy has been implicated in an increasing number of clinical scenarios, notably infectious diseases, cancer, neurodegenerative diseases, and autoimmunity. The recent identification of the importance of autophagy genes in the genetic susceptibility to Crohn ’ s disease suggests that a selective autophagic response may play a crucial role in the pathogenesis of common complex immune-mediated diseases. In this review, we discuss the autophagic mechanisms, their molecular regulation, and summarize their clinical relevance. This progress has led to great interest in the therapeutic potential of manipulation of both selective and nonselective autophagy in established disease. INTRODUCTION The ability to adapt to environmental change is essential for sur- Autophagy encompasses several distinct processes involving vival. This is true for the organism as a whole and for individual the delivery of portions of the cytoplasm to the lysosome for cells alike. -

Characterization of Gf a Drosophila Trimeric G Protein Alpha Subunit
Characterization of Gf a Drosophila trimeric G protein alpha subunit Naureen Quibria Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2012 © 2012 Naureen Quibria All rights reserved Abstract Characterization of Gf a Drosophila trimeric G-protein alpha subunit Naureen Quibria In the morphogenesis of tissue development, how coordination of patterning and growth achieve the correct organ size and shape is a principal question in biology. Efficient orchestrating mechanisms are required to achieve this and cells have developed sophisticated systems for reception and interpretation of the multitude of extracellular stimuli to which they are exposed. Plasma membrane receptors play a key role in the transmission of such signals. G-protein coupled receptors (GPCRs) are the largest class of cell surface receptors that respond to an enormous diversity of extracellular stimuli, and are critical mediators of cellular signal transduction in eukaryotic organisms. Signaling through GPCRs has been well characterized in many biological contexts. While they are a major class of signal transducers, there are not many defined instances where GPCRs have been implicated in the process of development to date. The Drosophila wing provides an ideal model system to elucidate and address the role of GPCRs in development, as its growth is regulated by a small number of conserved signaling pathways. In my thesis work, I address the role of a trimeric G alpha protein in Drosophila, Gαf, and what part it may play in development. In particular, I explore the role of Gαf as an alpha subunit of a trimeric complex, to determine what heptahelical receptors might act as its cognate receptor. -

Deficiency in Class III PI3-Kinase Confers Postnatal Lethality with IBD
ARTICLE DOI: 10.1038/s41467-018-05105-8 OPEN Deficiency in class III PI3-kinase confers postnatal lethality with IBD-like features in zebrafish Shaoyang Zhao1,2,3, Jianhong Xia2,3, Xiuhua Wu2,3, Leilei Zhang2,3, Pengtao Wang2,3, Haiyun Wang2,3, Heying Li2, Xiaoshan Wang 2, Yan Chen 2, Jean Agnetti2, Yinxiong Li 2, Duanqing Pei2,3 & Xiaodong Shu2,3 The class III PI3-kinase (PIK3C3) is an enzyme responsible for the generation of phospha- tidylinositol 3-phosphate (PI3P), a critical component of vesicular membrane. Here, we report 1234567890():,; that PIK3C3 deficiency in zebrafish results in intestinal injury and inflammation. In pik3c3 mutants, gut tube forms but fails to be maintained. Gene expression analysis reveals that barrier-function-related inflammatory bowel disease (IBD) susceptibility genes (e-cadherin, hnf4a, ttc7a) are suppressed, while inflammatory response genes are stimulated in the mutants. Histological analysis shows neutrophil infiltration into mutant intestinal epithelium and the clearance of gut microbiota. Yet, gut microorganisms appear dispensable as mutants cultured under germ-free condition have similar intestinal defects. Mechanistically, we show that PIK3C3 deficiency suppresses the formation of PI3P and disrupts the polarized dis- tribution of cell-junction proteins in intestinal epithelial cells. These results not only reveal a role of PIK3C3 in gut homeostasis, but also provide a zebrafish IBD model. 1 School of Life Sciences, University of Science and Technology of China, 230027 Hefei, Anhui, China. 2 CAS Key Laboratory of Regenerative Biology, Guangzhou Institute of Biomedicine and Health-Guangzhou Medical University Joint School of Biological Sciences, South China Institute for Stem Cell Biology and Regenerative Medicine, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, 510530 Guangzhou, China. -

Xxviiith Belgian Week of Gastroenterology 2016 All Abstracts Belgian Association for the Study of the Liver (BASL)
XXVIIIth Belgian Week of Gastroenterology 2016 All Abstracts Belgian Association for the Study of the Liver (BASL) A01 Light-to-moderate alcohol intake increases the risk of hepatocellular carcinoma in patients with HCV-related compensated cirrhosis: a prospective study H. VANDENBULCKE (1), C. MORENO (2), I. COLLE (3), J. KNEBEL (4), S. FRANCQUE (5), T. SERSTÉ (6), C. GEORGE (7), C. DE GALOCSY (8), W. LALEMAN (9), J. DELWAIDE (10), H. ORLENT (11), L. LASSER (12), E. TRÉPO (2), H. VAN VLIERBERGHE (3), P. MICHIELSEN (5), M. VAN GOSSUM (6), M. DE VOS (1), A. MAROT (13), C. DOERIG (13), M. ADLER (2), J. HENRION (1), P. DELTENRE (13) / [1] Hôpital de Jolimont, Haine-Saint-Paul, Belgium, Departement of Gastroenterology and Hepatology, [2] Erasme Hospital, Brussels, Belgium, Department of Gastroenterology, Hepatopancreatology and Digestive Oncology, [3] Ghent University, Ghent, Belgium, Departement of Gastroenterology and Hepatology, [4] Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland, Division of Radiology, [5] Antwerp University Hospital, Edegem, Belgium, Departement of Gastroenterology and Hepatology, [6] CHU Saint-Pierre, Brussels, Belgium, Departement of Gastroenterology and Hepatology, [7] AZ Groeninge, Kortrijk, Belgium, Departement of Gastroenterology and Hepatology, [8] Hôp. Iris Sud Bracops, Bruxelles, Belgium, Departement of Gastroenterology and Hepatology, [9] KU, Leuven, Belgium, Departement of Gastroenterology and Hepatology, [10] CHU Liege, Liège, Belgium, Departement of Gastroenterology and Hepatology, [11] AZ St. Jan Brugge AV, Brugge, Belgium, Departement of Gastroenterology and Hepatology, [12] CHU Brugmann, , Belgium, Departement of Gastroenterology and Hepatology, [13] Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland, Division of Gastroenterology and Hepatology Introduction: Whether light-to-moderate alcohol intake increases the risk of complications in patients with HCV-related cirrhosis remains unclear. -

Ifng-Induced Irgm1 Promotes Tumorigenesis of Melanoma Via Dual Regulation of Apoptosis and Bif-1-Dependent Autophagy
Oncogene (2015) 34, 5363–5371 © 2015 Macmillan Publishers Limited All rights reserved 0950-9232/15 www.nature.com/onc ORIGINAL ARTICLE IFNg-induced Irgm1 promotes tumorigenesis of melanoma via dual regulation of apoptosis and Bif-1-dependent autophagy H Dong1,2,5, L Tian1,5,RLi3,CPei1,YFu1, X Dong1, F Xia1, C Wang1,WLi1, X Guo1,CGu1,BLi1, A Liu4, H Ren1, C Wang2 and H Xu1 Interferon gamma (IFNg) has been known as the regulator for both tumor immune surveillance and tumorgenesis. However, mechanisms underlying the resistance of tumor cell to IFNg have yet been fully understood. In the current study, we showed that immunity-related GTPase family member 1 (mouse: Irgm1; human: IRGM) is essential for IFNg-mediated regulation of tumor cell growth in melanoma. IRGM/Irgm1 was highly expressed in human and mouse melanoma. IFNg and starvation synergistically induced Irgm1 expression in melanoma B16 cells. In vivo, injection of Irgm1-siRNA-treated cells significantly reduced the number of tumor nodules and prolonged the mice survival. In vitro, knockdown endogenous or IFNg-induced Irgm1 significantly decreases the proliferation and increases apoptosis of B16 cells. In addition, suppressing Irgm1 decreased the IFNg/starvation-induced autophagy, while overexpressing Irgm1 significantly increased autophagy and rescued starvation-challenged cells. Moreover, IFNg and starvation-induced the co-localization of Irgm1 with Bax-interacting factor 1 (Bif-1). Knockdown of Bif-1 decreased Irgm1-mediated tumor cell autophagy. Taken together, these data reveal an Irgm1-dependent mechanism that promotes the tumorigenesis of melanoma via dual regulation of apoptosis and Bif-1-dependent autophagy. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Interplay Between Cellular and Molecular Mechanisms Underlying Inflammatory Bowel Diseases Development—A Focus on Ulcerative Colitis
cells Review Interplay between Cellular and Molecular Mechanisms Underlying Inflammatory Bowel Diseases Development—A Focus on Ulcerative Colitis Iuliana Samoilă 1 , Sorina Dinescu 1,2,* and Marieta Costache 1,2 1 Department of Biochemistry and Molecular Biology, University of Bucharest, 050095 Bucharest, Romania; [email protected] (I.S.); [email protected] (M.C.) 2 Research Institute of the University of Bucharest, 050663 Bucharest, Romania * Correspondence: [email protected] Received: 16 May 2020; Accepted: 7 July 2020; Published: 9 July 2020 Abstract: Inflammatory bowel diseases (IBD) are defined by the continuous inflammation of the gastrointestinal tract. During inflammation, the number of pathogens in the intestinal epithelium increases, leading to inflammasome assembly. Inflammasome activation is meant to protect the intestinal epithelial barrier from further damage by maintaining homeostasis. Although its purpose is to protect the cells, excessive nucleotide-binding oligomerization domain-like receptor and pyrin domain-containing protein 3 (NLRP3) inflammasome assembly is responsible for the synthesis of a high number of pro-inflammatory cytokines. The activation of two crucial pathways, autophagy process, and unfolded protein response, is initiated for restoring homeostasis. Aberrant expression of miRNAs and lncRNAs also interfere with the pathogenic mechanisms of IBD, as these non-coding transcripts play key roles in regulation of biological processes, such as inflammation and immunity. This review thoroughly describes the cellular and molecular mechanism that trigger and perpetuate inflammation in ulcerative colitis (UC) patients. Keywords: inflammasome; NLRP3; autophagy; miRNAs in IBD; inflammatory bowel diseases 1. Introduction Inflammatory bowel diseases (IBD) are characterized by chronic inflammation of the gastrointestinal tract, with alternating phases of clinical relapse and remission [1]. -

Autophagy, Unfolded Protein Response, and Neuropilin-1 Cross-Talk in SARS-Cov-2 Infection: What Can Be Learned from Other Coronaviruses
International Journal of Molecular Sciences Review Autophagy, Unfolded Protein Response, and Neuropilin-1 Cross-Talk in SARS-CoV-2 Infection: What Can Be Learned from Other Coronaviruses Morvarid Siri 1 , Sanaz Dastghaib 2 , Mozhdeh Zamani 1 , Nasim Rahmani-Kukia 3 , Kiarash Roustai Geraylow 4 , Shima Fakher 3, Fatemeh Keshvarzi 3, Parvaneh Mehrbod 5 , Mazaher Ahmadi 6 , Pooneh Mokarram 1,3,* , Kevin M. Coombs 7 and Saeid Ghavami 1,8,9,* 1 Autophagy Research Center, Shiraz University of Medical Sciences, Shiraz 7134845794, Iran; [email protected] (M.S.); [email protected] (M.Z.) 2 Endocrinology and Metabolism Research Center, Shiraz University of Medical Sciences, Shiraz 7193635899, Iran; [email protected] 3 Department of Biochemistry, Shiraz University of Medical Sciences, Shiraz 7134845794, Iran; [email protected] (N.R.-K.); [email protected] (S.F.); [email protected] (F.K.) 4 Student Research Committee, Semnan University of Medical Sciences, Semnan 3514799422, Iran; [email protected] 5 Influenza and Respiratory Viruses Department, Pasteur Institute of Iran, Tehran 1316943551, Iran; [email protected] 6 Faculty of Chemistry, Bu-Ali Sina University, Hamedan 6517838695, Iran; [email protected] 7 Department of Medical Microbiology and Infectious Diseases, Rady Faculty of Health Sciences, Max Rady Citation: Siri, M.; Dastghaib, S.; College of Medicine, University of Manitoba, Winnipeg, MB R3E 0J9, Canada; [email protected] Zamani, M.; Rahmani-Kukia, N.; 8 Department of Human Anatomy and Cell Science, Rady Faculty of Health Sciences, Max Rady College Geraylow, K.R.; Fakher, S.; Keshvarzi, of Medicine, University of Manitoba, Winnipeg, MB R3E 0J9, Canada F.; Mehrbod, P.; Ahmadi, M.; 9 Faculty of Medicine, Katowice School of Technology, 40-555 Katowice, Poland Mokarram, P.; et al. -
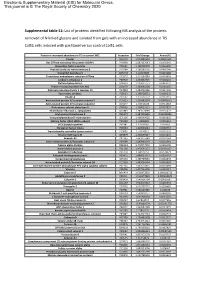
Supplemental Table S1
Electronic Supplementary Material (ESI) for Molecular Omics. This journal is © The Royal Society of Chemistry 2020 Supplemental table S1: List of proteins identified following MS analysis of the proteins removed of N-linked glycans and isolated from gels with an increased abundance in TIS Cal51 cells induced with paclitaxel versus control Cal51 cells. Protein in increased abundance in TIS vs control WCL Accession Fold Change Anova (P) Plectin Q15149 1.073855593 0.00691631 Ras GTPase-activating-like protein IQGAP1 P46940 1.087337643 0.0176342 Elongation factor1-gamma P26641 1.138709703 0.0116496 Peptidyl-prolyl cis-transisomerase B P23284 1.188383105 0.0436246 Dipeptidyl peptidase 3 Q9NY33 1.20163605 0.0215448 Transitional endoplasmic reticulum ATPase P55072 1.214194884 0.0449691 Carbonic anhydrase 2 P00918 1.232852325 0.0158141 Clathrin heavy chain 1 Q00610 1.239621773 0.0463237 Protein transport protein Sec 31A O94979 1.263565104 0.0284155 Aldo-ketoreductase family 1 member C1 Q04828 1.282092186 0.0324406 Spermidine synthase P19623 1.298728621 0.0196232 Plastin-3 P13797 1.310756772 0.0161319 Actin-related protein 2/3 complex subunit 5 O15511 1.333483524 0.00476923 Actin-related protein 2/3 complex subunit 2 O15144 1.35416168 0.0411018 Proteasome subunit alpha type-5 P28066 1.358015551 0.0337657 Thioredoxin reductase 1, cytoplasmic Q16881 1.383670089 0.0235472 Acyl-protein thioesterase 2 O95372 1.387415589 0.00233899 Isoaspartylpeptidase/L-asparaginase Q7L266 1.408149002 0.0319602 Splicing factor U2AF 65kDa subunit P26368 1.41489991 0.0256619 -

Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/Mtor Cascade Inhibitors: How Mutations Can Result in Therapy Resistance and How to Overcome Resistance
www.impactjournals.com/oncotarget/ Oncotarget, October, Vol.3, No 10 Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR Cascade Inhibitors: How Mutations Can Result in Therapy Resistance and How to Overcome Resistance James A. McCubrey1, Linda S. Steelman1, William H. Chappell1, Stephen L. Abrams1, Richard A. Franklin1, Giuseppe Montalto2, Melchiorre Cervello3, Massimo Libra4, Saverio Candido4, Grazia Malaponte4, Maria C. Mazzarino4, Paolo Fagone4, Ferdinando Nicoletti4, Jörg Bäsecke5, Sanja Mijatovic6, Danijela Maksimovic- Ivanic6, Michele Milella7, Agostino Tafuri8, Francesca Chiarini9, Camilla Evangelisti9, Lucio Cocco10, Alberto M. Martelli9,10 1 Department of Microbiology and Immunology, Brody School of Medicine at East Carolina University, Greenville, NC, USA 2 Department of Internal Medicine and Specialties, University of Palermo, Palermo, Italy 3 Consiglio Nazionale delle Ricerche, Istituto di Biomedicina e Immunologia Molecolare “Alberto Monroy”, Palermo, Italy 4 Department of Bio-Medical Sciences, University of Catania, Catania, Italy 5 Department of Medicine, University of Göttingen, Göttingen, Germany 6 Department of Immunology, Instititue for Biological Research “Sinisa Stankovic”, University of Belgrade, Belgrade, Serbia 7 Regina Elena National Cancer Institute, Rome, Italy 8 Sapienza, University of Rome, Department of Cellular Biotechnology and Hematology, Rome, Italy 9 Institute of Molecular Genetics, National Research Council-Rizzoli Orthopedic Institute, Bologna, Italy. 10 Department of Biomedical and Neuromotor Sciences, University of Bologna, Bologna, Italy Correspondence to: James A. McCubrey, email: [email protected] Keywords: Targeted Therapy, Therapy Resistance, Cancer Stem Cells, Raf, Akt, PI3K, mTOR Received: September 12, 2012, Accepted: October 18, 2012, Published: October 20, 2012 Copyright: © McCubrey et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. -

Melanomas Are Comprised of Multiple Biologically Distinct Categories
Melanomas are comprised of multiple biologically distinct categories, which differ in cell of origin, age of onset, clinical and histologic presentation, pattern of metastasis, ethnic distribution, causative role of UV radiation, predisposing germ line alterations, mutational processes, and patterns of somatic mutations. Neoplasms are initiated by gain of function mutations in one of several primary oncogenes, typically leading to benign melanocytic nevi with characteristic histologic features. The progression of nevi is restrained by multiple tumor suppressive mechanisms. Secondary genetic alterations override these barriers and promote intermediate or overtly malignant tumors along distinct progression trajectories. The current knowledge about pathogenesis, clinical, histological and genetic features of primary melanocytic neoplasms is reviewed and integrated into a taxonomic framework. THE MOLECULAR PATHOLOGY OF MELANOMA: AN INTEGRATED TAXONOMY OF MELANOCYTIC NEOPLASIA Boris C. Bastian Corresponding Author: Boris C. Bastian, M.D. Ph.D. Gerson & Barbara Bass Bakar Distinguished Professor of Cancer Biology Departments of Dermatology and Pathology University of California, San Francisco UCSF Cardiovascular Research Institute 555 Mission Bay Blvd South Box 3118, Room 252K San Francisco, CA 94158-9001 [email protected] Key words: Genetics Pathogenesis Classification Mutation Nevi Table of Contents Molecular pathogenesis of melanocytic neoplasia .................................................... 1 Classification of melanocytic neoplasms -

The Small Gtpase Rheb Affects Central Brain Neuronal Morphology and Memory Formation in Drosophila
The Small GTPase Rheb Affects Central Brain Neuronal Morphology and Memory Formation in Drosophila Heather L. D. Brown1¤a, Karla R. Kaun2¤b, Bruce A. Edgar1*¤c 1 Basic Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, Washington, United States of America, 2 Department of Anatomy, University of California San Francisco, San Francisco, California, United States of America Abstract Mutations in either of two tumor suppressor genes, TSC1 or TSC2, cause tuberous sclerosis complex (TSC), a syndrome resulting in benign hamartomatous tumors and neurological disorders. Cellular growth defects and neuronal disorganization associated with TSC are believed to be due to upregulated TOR signaling. We overexpressed Rheb, an upstream regulator of TOR, in two different subsets of D. melanogaster central brain neurons in order to upregulate the Tsc- Rheb-TOR pathway. Overexpression of Rheb in either the mushroom bodies or the insulin producing cells resulted in enlarged axon projections and cell bodies, which continued to increase in size with prolonged Rheb expression as the animals aged. Additionally, Rheb overexpression in the mushroom bodies resulted in deficiencies in 3 hr but not immediate appetitive memory. Thus, Rheb overexpression in the central brain neurons of flies causes not only morphological phenotypes, but behavioral and aging phenotypes that may mirror symptoms of TSC. Citation: Brown HLD, Kaun KR, Edgar BA (2012) The Small GTPase Rheb Affects Central Brain Neuronal Morphology and Memory Formation in Drosophila. PLoS ONE 7(9): e44888. doi:10.1371/journal.pone.0044888 Editor: Shree Ram Singh, National Cancer Institute, United States of America Received February 28, 2012; Accepted August 14, 2012; Published September 19, 2012 Copyright: ß 2012 Brown et al.