Isolation and Molecular Characterization
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Charter of Independence: a Critical Study of Mujib's Six Point Programme
Ayyaz GullI CHARTER OF INDEPENDENCE: A CRITICAL STUDY OF MUJIB'S SIX POINT PROGRAMME Abstract This paper will try to explore the role of Bengali political leadership to transform the dream of separation of East Pakistan into reality. It will also provide a detailed and systematic study of Six Point Programme of Awami League which proved to be a 'charter of independence' and gave a comprehensive analysis of the basic demands of East Pakistanis and successfully combined public opinions in order to get mass support in the struggle for freedom from the West Pakistan. Moreover, this study will seek several waves of criticism regarding Six Point Programme by the state of Pakistan, political parties of West and East Pakistan, and even by the people within the Awami League. Key Words: Six Point Formula, Two Economy Thesis, Secessionist Movement, Economic Disparities, Conspiracy Theories The East Bengalis political elite played an important role in the separation of East Pakistan. It was economic exploitation which gave them an ample opportunity to win over popular support. They were conscious of these distinct geographical and cultural features, and they lost no occasion to project the differences between the two wings. They highlighted the points of ‘separateness’ in their speeches in the Constituent Assembly and the Provincial Assemblies. For instance, Abdul Mansur Ahmad, a prominent member from East Pakistan, observed in Constituent Assembly Pakistan is a unique country having two wings which are separated by a distance of more than a thousand miles…religion and common struggle are the only common factors… with the exception of these two things, all other factors, viz the language, the culture…practically everything is different. -

IWCSS 2020 Organizing and Program Committees
IWCSS 2020 Organizing and Program Committees General Chairs J. Christopher Westland, University of Illinois (Chicago), USA Hai Jiang, Arkansas State University, USA Aniello Castiglione, University of Naples Parthenope, Italy Program Chairs Valentina Emilia Balas, "Aurel Vlaicu" University of Arad, Romania Yang Xu, Hunan University, China Program Committee Md Atiqur Rahman Ahad, University of Dhaka, Bangladesh Saqib Ali, University of Agriculture Faisalabad, Pakistan Flora Amato, University of "Naples Federico II", Italy Ranbir Singh Batth, Lovely Professional University, India Oana Geman, Stefan cel Mare University of Suceava, Romania Che-Lun Hung, Providence University, Taiwan Xing Gao, University of Memphis, USA Donghoon Kim, Arkansas State University, USA Gabor Kiss, Obuda University, Hungary Konstantinos Kolias, University of Idaho, USA Neeraj Kumar, Thapar Institute of Engineering and Technology, India Anyi Liu, Oakland University, USA Weizhi Meng, Technical University of Denmark, Denmark Fabio Narducci, University of Naples "Parthenope", Italy Anand Nayyar, Duytan University, Vietnam Reza Meimandi Parizi, Kennesaw State University, USA Emil Pricop, Petroleum - Gas University of Ploiesti, Romania Quan Qian, Shanghai University, China Vijender Kr. Solanki, CMR Institute of Technology, Hyderabad, India Zhiyuan Tan, Napier University, UK Bing Tang, Hunan University of Science and Technology, China Muhhamad Imran Tariq, Superior University Lahore, Pakistan Ning Wang, Rowan University, USA Haitao Xu, Arizona State University, USA Pengcheng You, Johns Hopkins University, USA Ji Zhang, University of Southern Queensland, Australia Philippe Fournier-Viger, Harbin Institute of Technology, China Hairong Lv, Tsinghua University, China Shuo Feng, McMaster University, Canada Venkatasamy Sureshkumar, PSG College of Technology, India Qiao Hu, Hunan University, China Tasmina Islam, University of Kent, UK lxi Xiaokang Wang, St. -

Men's Athlete Profiles 1 49KG – SIMPLICE FOTSALA – CAMEROON
Gold Coast 2018 Commonwealth Games - Men's Athlete Profiles 49KG – SIMPLICE FOTSALA – CAMEROON (CMR) Date Of Birth : 09/05/1989 Place Of Birth : Yaoundé Height : 160cm Residence : Region du Centre 2018 – Indian Open Boxing Tournament (New Delhi, IND) 5th place – 49KG Lost to Amit Panghal (IND) 5:0 in the quarter-final; Won against Muhammad Fuad Bin Mohamed Redzuan (MAS) 5:0 in the first preliminary round 2017 – AFBC African Confederation Boxing Championships (Brazzaville, CGO) 2nd place – 49KG Lost to Matias Hamunyela (NAM) 5:0 in the final; Won against Mohamed Yassine Touareg (ALG) 5:0 in the semi- final; Won against Said Bounkoult (MAR) 3:1 in the quarter-final 2016 – Rio 2016 Olympic Games (Rio de Janeiro, BRA) participant – 49KG Lost to Galal Yafai (ENG) 3:0 in the first preliminary round 2016 – Nikolay Manger Memorial Tournament (Kherson, UKR) 2nd place – 49KG Lost to Ievgen Ovsiannikov (UKR) 2:1 in the final 2016 – AIBA African Olympic Qualification Event (Yaoundé, CMR) 1st place – 49KG Won against Matias Hamunyela (NAM) WO in the final; Won against Peter Mungai Warui (KEN) 2:1 in the semi-final; Won against Zoheir Toudjine (ALG) 3:0 in the quarter-final; Won against David De Pina (CPV) 3:0 in the first preliminary round 2015 – African Zone 3 Championships (Libreville, GAB) 2nd place – 49KG Lost to Marcus Edou Ngoua (GAB) 3:0 in the final 2014 – Dixiades Games (Yaounde, CMR) 3rd place – 49KG Lost to Marcus Edou Ngoua (GAB) 3:0 in the semi- final 2014 – Cameroon Regional Tournament 1st place – 49KG Won against Tchouta Bianda (CMR) -

News Letter Embassy of Pakistan Jakarta April - June 2015
NEWS LETTER EMBASSY OF PAKISTAN JAKARTA APRIL - JUNE 2015 Contents of the Issue Death of Indonesian Ambassador to Pakistan, Burhan Muhammad · Death of Indonesian and His Wife in Pakistan Helicopter Crash Ambassador to Pakistan Burhan Muhammad 1 · Asian-African Conference 2015 2 · Media Engagement 2 · Indonesia Participation in My Karachi Expo 2 Tributes paid to Ambassador Burhan Muhammad at · Indonesia-Pakistan Gedung Pancasila, Jakarta Bilateral Data 2 Indonesian ambassador to Pakistan President of Indonesia, Joko Widodo · Asian-African Business Burhan Muhammad died at the expressed deep sorrow on the passing Summit 2015 3 Singapore General Hospital on 19th of Indonesian Ambassador to May 2015, 11 days after the Pakistan, Burhan Muhammad. · Visit of NDU Delegates 3 Pakistani military helicopter he was The body was taken to the family · Pakistan Stall at AITIS 2015 3 travelling on crashed in the home where he was buried with full mountains of Gilgit-Baltistan, military honours. Pakistan High · Economic Activities 3 Pakistan on May 8. The accident Commissioner to Singapore H.E. · Obituary of Ambassador killed seven people on the spot, Tanveer Khaskheli and Pakistan including his wife Heri Listyawati Charge d’ Affairs to Indonesia, Syed Burhan Muhammad 4 and envoys from the Philippines and Zahid Raza, represented government · Activities at Norway. The envoys were part of a of Pakistan at the funeral. Pakistan Embassy, Jakarta 4 large group of foreign dignitaries Earlier, Minister of Defence being ferried to the inauguration of a Production, Mr. Rana Tanveer Cd'A / Counsellor ski resort chairlift in the town of Hussein brought the dead body of Syed Zahid Raza Naltar. -

Partition of Punjab: Sikhs and Lyallpur, Explores the Significance of Lyallpur in Partition of Punjab
THIS DISSERTATION IS BEING SUBMITTED TO THE UNIVERSITY OF THE PUNJAB IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY IN HISTORY ROLE OF MIGRANTS IN MAKING OF MODERN FAISALABAD 1947-1960 Submitted By: Muhammad Abrar Ahmad Ph. D. 2011-2016 Research Supervisor: Prof. Dr. Muhammad Iqbal Chawla Dean, Faculty of Arts & Humanities University of the Punjab Department of History & Pakistan Studies University of the Punjab, Lahore 2016 In the name of ALLAH, the creator of the universe the most beneficent, most merciful. DECLARATION I, hereby, declare that this Ph. D thesis titled “Role of Migrants in Making of Modern Faisalabad 1947-1960” is the result of my personal research and is not being submitted concurrently to any other University for any degree or whatsoever. Muhammad Abrar Ahmad Ph. D. Scholar I CERTIFICATE Certificate by Research Supervisor This is to certify that Muhammad Abrar Ahmad has completed the Dissertation entitled “Role of Migrants in Making of Modern Faisalabad 1947-1960” under my supervision. It fulfills the requirements necessary for submission of the dissertation for the Doctor of Philosophy in History. Supervisor Prof. Dr. Muhammad Iqbal Chawla Dean, Faculty of Arts & Humanities, University of the Punjab, Lahore. Submitted Through Prof. Dr. Muhammad Iqbal Chawla Chairman, Department of History & Pakistan Studies, University of the Punjab, Lahore External Examiner II DEDICATED To My Loving Amma Gee (Late) who has been a constant source of warmth and inspiration III Acknowledgement First of all, I offer my bow of humility to Almighty Allah who opened my mind and enabled me to carry out this noble task of contributing few drops in ocean of knowledge. -

Supplemental Statement Washington, DC 20530 Pursuant to the Foreign Agents Registration Act of 1938, As Amended
Received by NSD/FARA Registration Unit 07/17/2013 12:53:25 PM OMB NO. 1124-0002; Expires February 28, 2014 «JJ.S. Department of Justice Supplemental Statement Washington, DC 20530 Pursuant to the Foreign Agents Registration Act of 1938, as amended For Six Month Period Ending 06/30/2013 (Insert date) I - REGISTRANT 1. (a) Name of Registrant (b) Registration No. Pakistan Tehreek e Insaf 5975 (c) Business Address(es) of Registrant 315 Maple street Richardson TX, 75081 Has there been a change in the information previously furnished in connection with the following? (a) If an individual: (1) Residence address(es) Yes Q No D (2) Citizenship Yes Q No Q (3) Occupation Yes • No D (b) If an organization: (1) Name Yes Q No H (2) Ownership or control Yes • No |x] - (3) Branch offices Yes D No 0 (c) Explain fully all changes, if any, indicated in Items (a) and (b) above. IF THE REGISTRANT IS AN INDIVIDUAL, OMIT RESPONSE TO ITEMS 3,4, AND 5(a). 3. If you have previously filed Exhibit C1, state whether any changes therein have occurred during this 6 month reporting period. Yes D No H If yes, have you filed an amendment to the Exhibit C? Yes • No D If no, please attach the required amendment. I The Exhibit C, for which no printed form is provided, consists of a true copy of the charter, articles of incorporation, association, and by laws of a registrant that is an organization. (A waiver of the requirement to file an Exhibit C may be obtained for good cause upon written application to the Assistant Attorney General, National Security Division, U.S. -
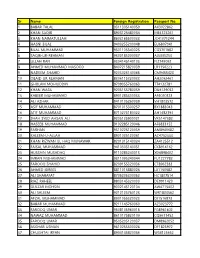
Sr Name Foreign Registration Passport No. 1 BABAR
Sr Name Foreign Registration Passport No. -

List of Shareholder-Dividend Withheld
Ghandhara Industries Limited List of shareholders - Dividend Withheld MEMBER NET SR # SHAREHOLDER'S NAME ADDRESS FOLIO PAR ID PAYABLE 1 00002-000 MISBAHUDDIN FAROOQI D-151 BLOCK-B NORTH NAZIMABADKARACHI. 461 2 00004-000 FARHAT AFZA BEGUM MST. 166 ABU BAKR BLOCK NEW GARDEN TOWN,LAHORE 1,302 3 00005-000 ALLAH RAKHA 135-MUMTAZ STREETGARHI SHAHOO,LAHORE. 3,293 4 00006-000 AKHTAR A. PERVEZ C/O. AMERICAN EXPRESS CO.87 THE MALL, LAHORE. 180 5 00008-000 DOST MUHAMMAD C/O. NATIONAL MOTORS LIMITEDHUB CHAUKI ROAD, S.I.T.E.KARACHI. 46 6 00010-000 JOSEPH F.P. MASCARENHAS MISQUITA GARDEN CATHOLIC COOP.HOUSING SOCIETY LIMITED BLOCK NO.A-3,OFF: RANDLE ROAD KARACHI. 1,544 7 00013-000 JOHN ANTHONY FERNANDES A-6, ANTHONIAN APT. NO. 1,ADAM ROAD,KARACHI. 5,004 8 00014-000 RIAZ UL HAQ HAMID C-103, BLOCK-XI, FEDERAL.B.AREAKARACHI. 69 9 00015-000 SAIED AHMAD SHAIKH C/O.COMMODORE (RETD) M. RAZI AHMED71/II, KHAYABAN-E-BAHRIA, PHASE-VD.H.A. KARACHI-46. 214 10 00016-000 GHULAM QAMAR KURD 292/8, AZIZABAD FEDERAL B. AREA,KARACHI. 129 11 00017-000 MUHAMMAD QAMAR HUSSAIN C/O.NATIONAL MOTORS LTD.HUB CHAUKI ROAD, S.I.T.E.,P.O.BOX-2706, KARACHI. 218 12 00018-000 AZMAT NAWAZISH AZMAT MOTORS LIMITED, SC-43,CHANDNI CHOWK, STADIUM ROAD,KARACHI. 1,585 13 00021-000 MIRZA HUSSAIN KASHANI HOUSE NO.R-1083/16,FEDERAL B. AREA, KARACHI 434 14 00023-000 RAHAT HUSSAIN PLOT NO.R-483, SECTOR 14-B,SHADMAN TOWN NO.2, NORTH KARACHI,KARACHI. -

HCAR CDC LIST AS on MARCH 21, 2018 Sr No Member Folio Parid
HCAR CDC LIST AS ON MARCH 21, 2018 Sr No Member Folio ParID CDC A/C # SHARE HOLDER'S NAME ADDRESS 1 CDC-1 00208 30 ALFA ADHI SECURITIES (PVT) LTD. SUITE NO. 303, 3RD FLOOR, MUHAMMAD BIN QASIM ROAD, I.I. CHUNDRIGAR ROAD KARACHI 2 CDC-2 00208 3075 Rafiq III-F, 13/7, NAZIMABAD NO. 3, 74600 KARACHI 3 CDC-3 00208 3729 Omar Farooq HOUSE NO B-144, BLOCK-3, GULISTAN-E-JOUHAR KARACHI 4 CDC-4 00208 7753 AFSHAN SHAKIL A-82, S.B.C.H.S. BLOCK 12, GULISTAN-E-JAUHAR KARACHI 5 CDC-5 00208 8827 SARWAR DIN L-361, SHEERIN JINNAH COLONY , CLIFTON KARACHI 6 CDC-6 00208 11805 SHUNIL KUMAR OFFICE # 105,HUSSAIN TRADE CENTER, ALTAF HUSSAIN ROAD, NEW CHALI KARACHI 7 CDC-7 00208 12126 ABDUL SAMAD D-9, DAWOOD COLONY, SHARFABAD, NEAR T.V. STATION. KARACHI 8 CDC-8 00208 18206 Syeda Sharmeen Flat # 506, Ana Classic Apartment, Ghulam Hussain Qasim Road, Garden West KARACHI 9 CDC-9 00208 18305 Saima Asif FLAT NO: 306, MOTIWALA APARTMENT, PLOT NO: BR 5/16, MULJI STREET, KHARADHAR KARACHI 10 CDC-10 00208 18511 Farida FLAT NO: 306, MOTIWALA APARTMENT, PLOT NO: BR 5/16, MULJI STREET, KHARADHAR, KARACHI 11 CDC-11 00208 19626 MANOHAR LAL MANOHAR EYE CLINIC JACOBABAD 12 CDC-12 00208 21945 ADNAN FLAT NO 101, PLOT NO 329, SIDRA APPARTMENT, GARDEN EAST, BRITTO ROAD, KARACHI 13 CDC-13 00208 25672 TAYYAB IQBAL FLAT NO. 301, QASR-E-GUL, NEAR T.V. STATION, JAMAL-UD-DIN AFGHANI ROAD, SHARFABAD KARACHI 14 CDC-14 00208 26357 MUHAMMAD SALEEM KHAN G 26\9,LIAQUAT SQUARE, MALIR COLONY, KARACHI 15 CDC-15 00208 27165 SABEEN IRFAN 401,HEAVEN PRIDE,BLOCK-D, FATIMA JINNAH COLONY,JAMSHED ROAD#2, KARACHI 16 CDC-16 00208 27967 SANA UL HAQ 3-F-18-4, NAZIMABAD KARACHI 17 CDC-17 00208 29112 KHALID FAREED SONIA SQUARE, FLAT # 05, STADIUM ROAD, CHANDNI CHOWK, KARACHI 5 KARACHI 18 CDC-18 00208 29369 MOHAMMAD ZEESHAN AHMED FLAT NO A-401, 4TH FLOOR, M.L GARDEN PLOT NO, 690, JAMSHED QUARTERS, KARACHI 19 CDC-19 00208 30151 SANIYA MUHAMMAD RIZWAN HOUSE # 4, PLOT # 3, STREET# 02, MUSLIMABAD, NEAR DAWOOD ENGG. -

Unclaimed Deposit 2014
Details of the Branch DETAILS OF THE DEPOSITOR/BENEFICIARIYOF THE INSTRUMANT NAME AND ADDRESS OF DEPOSITORS DETAILS OF THE ACCOUNT DETAILS OF THE INSTRUMENT Transaction Federal/P rovincial Last date of Name of Province (FED/PR deposit or in which account Instrume O) Rate Account Type Currency Rate FCS Rate of withdrawal opened/instrume Name of the nt Type In case of applied Amount Eqv.PKR Nature of Deposit ( e.g Current, (USD,EUR,G Type Contract PKR (DD-MON- Code Name nt payable CNIC No/ Passport No Name Address Account Number applicant/ (DD,PO, Instrument NO Date of issue instrumen date Outstandi surrender (LCY,UFZ,FZ) Saving, Fixed BP,AED,JPY, (MTM,FC No (if conversio YYYY) Purchaser FDD,TDR t (DD-MON- ng ed or any other) CHF) SR) any) n , CO) favouring YYYY) the Governm ent 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 PRIX 1 Main Branch Lahore PB Dir.Livestock Quetta MULTAN ROAD, LAHORE. 54500 LCY 02011425198 CD-MISC PHARMACEUTICA TDR 0000000189 06-Jun-04 PKR 500 12-Dec-04 M/S 1 Main Branch Lahore PB MOHAMMAD YUSUF / 1057-01 LCY CD-MISC PKR 34000 22-Mar-04 1 Main Branch Lahore PB BHATTI EXPORT (PVT) LTD M/S BHATTI EXPORT (PVT) LTD M/SLAHORE LCY 2011423493 CURR PKR 1184.74 10-Apr-04 1 Main Branch Lahore PB ABDUL RAHMAN QURESHI MR ABDUL RAHMAN QURESHI MR LCY 2011426340 CURR PKR 156 04-Jan-04 1 Main Branch Lahore PB HAZARA MINERAL & CRUSHING IND HAZARA MINERAL & CRUSHING INDSTREET NO.3LAHORE LCY 2011431603 CURR PKR 2764.85 30-Dec-04 "WORLD TRADE MANAGEMENT M/SSUNSET LANE 1 Main Branch Lahore PB WORLD TRADE MANAGEMENT M/S LCY 2011455219 CURR PKR 75 19-Mar-04 NO.4,PHASE 11 EXTENTION D.H.A KARACHI " "BASFA INDUSTRIES (PVT) LTD.FEROZE PUR 1 Main Branch Lahore PB 0301754-7 BASFA INDUSTRIES (PVT) LTD. -

ISLAMABAD, MONDAY, OCTOBER 28, 2019 PART II Statutory
PART II] THE GAZETTE OF PAKISTAN, EXTRA., OCT., 28, 2019 2379 ISLAMABAD, MONDAY, OCTOBER 28, 2019 PART II Statutory Notifications (S. R. O.) GOVERNMENT OF PAKISTAN MINISTRY OF FEDERAL EDUCATION & PROFESSIONAL TRAINING (National Commission for Human Development) NOTIFICATION Islamabad, the 1st August, 2019 S. R. O. 1279(I)/2019.—In pursuance of the orders of Honourable Islamabad High Court, Islamabad in Writ Petitions No. 724/2015, 231/2015, 726/2015, 1071/2015, 1073/2015 dated 09-12-2015, W.P 1337, 2329/2017 dated 14-11-2017 and Crl.Org No.166-W/ 2016 followed by creation of 2641 posts of NCHD employees by Finance Division FA’s Organization’s vide U.O No.1387 / DFA (FE&PT) dated 21.06.2018 and subsequently notified by M/o Federal Education & Professional Training vide F.No. 1-16/2011-Admin dated 09-07-2018, the services of the following Officers/Officials of National Commission for Human Development (NCHD) are regularized on a revised grades at the initial of Basic Pay Scale with effect from 01-07-2013. (9732) Price: Rs. 5.00 [1665 (2019)/Ex. Gaz.] 2380 THE GAZETTE OF PAKISTAN, EXTRA., OCT., 28, 2019 [PART II SL. EC Employee Name Designation BPS CNIC Location Province 1 4889 Uzma Fida Deputy Director 18 33100-7412524-8 Jhang Punjab 2 4906 Ghulam Moinuddin Deputy Director 18 54400-6030445-5 Noshki Balochistan 3 5189 Syed Akbar Ali Shah Deputy Director 18 71302-6090438-9 Head Office Islamabad 4 4719 Ghulam Farooq Assistant Director 17 51602-6308331-9 Mastung Balochistan 5 5138 Muhammad Daud Assistant Director 17 55301-2045686-5 Quetta Balochistan -
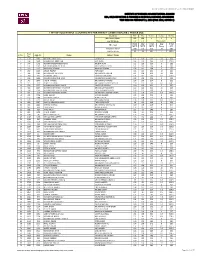
List of Successful Candidates for Direct
LIST OF SUCCESSFUL CANDIDATES ‐ FALL 2018, ROUND 1 INSTITUTE OF BUSINESS ADMINISTRATION, KARACHI BBA, BS (ACCOUNTING & FINANCE) & BS (SOCIAL SCIENCES) ADMISSIONS TEST HELD ON FEBRUARY 11, 2018 (FALL 2018, ROUND 1) LIST OF SUCCESSFUL CANDIDATES FOR DIRECT ADMISSION (BBA PROGRAM) SAT Test Math Eng Reading Analysis Writing Maximum Marks 800 800 8 8 8 Cut-Off Marks 670 650 Total of 18+ Math Eng Total Eng Total IBA Test MCQ MCQ MCQ Essay ALL Maximum Marks 200 176 376 20 396 Cut-Off Marks 96 92 204 7 224.5 Seat S. No. App No. Name Father's Name No. 1 130 1897 M FAAYEZ ZAKIR SATTI ZAKIR SATTI 124 100 224 10 234 2 146 1485 MUHAMMAD ABDULLAH ASIF JAVED 164 100 264 9.5 273.5 3 147 1172 MUHAMMAD EBRAHEEM ALAM MUSHIR ALAM 112 116 228 8 236 4 151 1481 SYED MIR MOMIN ALI SHAH SYED FAYYAZ ALI SHAH 132 104 236 8 244 5 249 2937 MAHRUKH REHMAN KAMRAN REHMAN 156 112 268 9 277 6 280 1649 ABDUR RAAFAY TARIQ AZIZ 104 120 224 10 234 7 299 1381 MUHAMMAD AHTISHAM MUHAMMAD ARSHAD 124 108 232 10 242 8 300 469 JAHANZAIB ZAKIR ZAKIR HUSSAIN KHAN 104 112 216 8.5 224.5 9 306 2450 MUHAMMAD JUNAID MIAN MUHAMMAD SHAHID MIAN 108 108 216 9 225 10 345 3006 HASAN FAROOQ MUHAMMAD FAROOQ 124 100 224 8 232 11 515 203 AHMED ZEESHAN MUHAMMAD SAJJAD MOGHAL 140 120 260 8.5 268.5 12 598 1507 MUHAMMAD UMAIR SHAFIQ SHAFIQ UR REHMAN 120 100 220 7 227 13 658 2857 MUHAMMAD HANNAN NAUSHAHI UBAIDULLAH NAUSHAHI 112 112 224 11 235 14 668 2649 MUHAMMAAD SAMEER KHAN FAZAL MAHMOOD KHAN 128 120 248 8 256 15 671 317 MUHAMMAD HASAN ARSHAD MUHAMMAD ARSHAD NAVEED 132 104 236 8.5 244.5 16 673 2726