Further Analysis and a Reply
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
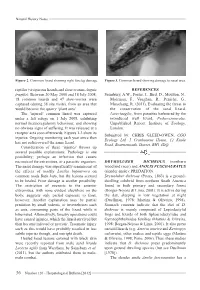
Reptiles (Viviparous Lizards and Slow-Worms Anguis REFERENCES Fragilis)
Natural History Notes Figure 2. Common lizard showing right foreleg damage. Figure 3. Common lizard showing damage to nasal area. reptiles (viviparous lizards and slow-worms Anguis REFERENCES fragilis). Between 20 May 2008 and 18 July 2008, Sainsbury, A.W., Foster, J., Bird, D., Moulton, N., 18 common lizards and 47 slow-worms were Molenaar, F., Vaughan, R., Peniche, G., captured (during 38 site visits), from an area that Marschang, R. (2011). Evaluating the threat to would become the quarry ‘plant area’. the conservation of the sand lizard, The ‘injured’ common lizard was captured Lacertaagilis, from parasites harboured by the under a felt refuge on 1 July 2008, exhibiting introduced wall lizard, Podarcismuralis. normal thermoregulatory behaviour, and showing Unpublished Report. Institute of Zoology, no obvious signs of suffering. It was released at a London. receptor area soon afterwards. Figures 1-3 show its Submitted by: CHRIS GLEED-OWEN, CGO injuries. Ongoing monitoring each year since then Ecology Ltd, 5 Cranbourne House, 12 Knole has not rediscovered the same lizard. Road, Bournemouth, Dorset, BH1 4DQ. Consideration of these ‘injuries’ throws up several possible explanations. Pathology is one possibility; perhaps an infection that causes necrosis of the extremities, or a parasitic organism. DRYMOLUBER DICHROUS (northern The nasal damage was superficially reminiscent of woodland racer) and ANOLIS FUSCOAURATUS the effects of toadfly Lucilia bufonivora on (slender anole): PREDATION. common toads Bufo bufo, but the lesions seemed Drymoluber dichrous (Peters, 1863) is a ground- to be healed. Frost damage is another possibility. dwelling colubrid from northern South America The restriction of necrosis to the anterior found in both primary and secondary forest extremities, with none evident elsewhere on the (Borges-Nojosa & Lima, 2001). -

Other Contributions
Other Contributions NATURE NOTES Amphibia: Caudata Ambystoma ordinarium. Predation by a Black-necked Gartersnake (Thamnophis cyrtopsis). The Michoacán Stream Salamander (Ambystoma ordinarium) is a facultatively paedomorphic ambystomatid species. Paedomorphic adults and larvae are found in montane streams, while metamorphic adults are terrestrial, remaining near natal streams (Ruiz-Martínez et al., 2014). Streams inhabited by this species are immersed in pine, pine-oak, and fir for- ests in the central part of the Trans-Mexican Volcanic Belt (Luna-Vega et al., 2007). All known localities where A. ordinarium has been recorded are situated between the vicinity of Lake Patzcuaro in the north-central portion of the state of Michoacan and Tianguistenco in the western part of the state of México (Ruiz-Martínez et al., 2014). This species is considered Endangered by the IUCN (IUCN, 2015), is protected by the government of Mexico, under the category Pr (special protection) (AmphibiaWeb; accessed 1April 2016), and Wilson et al. (2013) scored it at the upper end of the medium vulnerability level. Data available on the life history and biology of A. ordinarium is restricted to the species description (Taylor, 1940), distribution (Shaffer, 1984; Anderson and Worthington, 1971), diet composition (Alvarado-Díaz et al., 2002), phylogeny (Weisrock et al., 2006) and the effect of habitat quality on diet diversity (Ruiz-Martínez et al., 2014). We did not find predation records on this species in the literature, and in this note we present information on a predation attack on an adult neotenic A. ordinarium by a Thamnophis cyrtopsis. On 13 July 2010 at 1300 h, while conducting an ecological study of A. -

Xenosaurus Tzacualtipantecus. the Zacualtipán Knob-Scaled Lizard Is Endemic to the Sierra Madre Oriental of Eastern Mexico
Xenosaurus tzacualtipantecus. The Zacualtipán knob-scaled lizard is endemic to the Sierra Madre Oriental of eastern Mexico. This medium-large lizard (female holotype measures 188 mm in total length) is known only from the vicinity of the type locality in eastern Hidalgo, at an elevation of 1,900 m in pine-oak forest, and a nearby locality at 2,000 m in northern Veracruz (Woolrich- Piña and Smith 2012). Xenosaurus tzacualtipantecus is thought to belong to the northern clade of the genus, which also contains X. newmanorum and X. platyceps (Bhullar 2011). As with its congeners, X. tzacualtipantecus is an inhabitant of crevices in limestone rocks. This species consumes beetles and lepidopteran larvae and gives birth to living young. The habitat of this lizard in the vicinity of the type locality is being deforested, and people in nearby towns have created an open garbage dump in this area. We determined its EVS as 17, in the middle of the high vulnerability category (see text for explanation), and its status by the IUCN and SEMAR- NAT presently are undetermined. This newly described endemic species is one of nine known species in the monogeneric family Xenosauridae, which is endemic to northern Mesoamerica (Mexico from Tamaulipas to Chiapas and into the montane portions of Alta Verapaz, Guatemala). All but one of these nine species is endemic to Mexico. Photo by Christian Berriozabal-Islas. amphibian-reptile-conservation.org 01 June 2013 | Volume 7 | Number 1 | e61 Copyright: © 2013 Wilson et al. This is an open-access article distributed under the terms of the Creative Com- mons Attribution–NonCommercial–NoDerivs 3.0 Unported License, which permits unrestricted use for non-com- Amphibian & Reptile Conservation 7(1): 1–47. -

Zootaxa, Molecular Phylogeny, Classification, and Biogeography Of
Zootaxa 2067: 1–28 (2009) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2009 · Magnolia Press ISSN 1175-5334 (online edition) Molecular phylogeny, classification, and biogeography of West Indian racer snakes of the Tribe Alsophiini (Squamata, Dipsadidae, Xenodontinae) S. BLAIR HEDGES1, ARNAUD COULOUX2, & NICOLAS VIDAL3,4 1Department of Biology, 208 Mueller Lab, Pennsylvania State University, University Park, PA 16802-5301 USA. E-mail: [email protected] 2Genoscope. Centre National de Séquençage, 2 rue Gaston Crémieux, CP5706, 91057 Evry Cedex, France www.genoscope.fr 3UMR 7138, Département Systématique et Evolution, Muséum National d’Histoire Naturelle, CP 26, 57 rue Cuvier, 75005 Paris, France 4Corresponding author. E-mail : [email protected] Abstract Most West Indian snakes of the family Dipsadidae belong to the Subfamily Xenodontinae and Tribe Alsophiini. As recognized here, alsophiine snakes are exclusively West Indian and comprise 43 species distributed throughout the region. These snakes are slender and typically fast-moving (active foraging), diurnal species often called racers. For the last four decades, their classification into six genera was based on a study utilizing hemipenial and external morphology and which concluded that their biogeographic history involved multiple colonizations from the mainland. Although subsequent studies have mostly disagreed with that phylogeny and taxonomy, no major changes in the classification have been proposed until now. Here we present a DNA sequence analysis of five mitochondrial genes and one nuclear gene in 35 species and subspecies of alsophiines. Our results are more consistent with geography than previous classifications based on morphology, and support a reclassification of the species of alsophiines into seven named and three new genera: Alsophis Fitzinger (Lesser Antilles), Arrhyton Günther (Cuba), Borikenophis Hedges & Vidal gen. -

A Morphological and Molecular Study of Hydrodynastes Gigas (Serpentes, Dipsadidae), a Widespread Species from South America
A morphological and molecular study of Hydrodynastes gigas (Serpentes, Dipsadidae), a widespread species from South America Priscila S. Carvalho1,2, Hussam Zaher3, Nelson J. da Silva Jr4 and Diego J. Santana1 1 Instituto de Biociências, Universidade Federal de Mato Grosso do Sul, Campo Grande, Mato Grosso do Sul, Brazil 2 Instituto de Biociências, Letras e Ciências Exatas, Universidade Estadual Paulista, São José do Rio preto, São Paulo, Brazil 3 Museu de Zoologia da Universidade de São Paulo, São Paulo, São Paulo, Brazil 4 Escola de Ciências Médicas, Farmacêuticas e Biomédicas, Pontifícia Universidade Católica de Goiás, Goiânia, Goiás, Brazil ABSTRACT Background. Studies with integrative approaches (based on different lines of evidence) are fundamental for understanding the diversity of organisms. Different data sources can improve the understanding of the taxonomy and evolution of snakes. We used this integrative approach to verify the taxonomic status of Hydrodynastes gigas (Duméril, Bibron & Duméril, 1854), given its wide distribution throughout South America, including the validity of the recently described Hydrodynastes melanogigas Franco, Fernandes & Bentim, 2007. Methods. We performed a phylogenetic analysis of Bayesian Inference with mtDNA 16S and Cytb, and nuDNA Cmos and NT3 concatenated (1,902 bp). In addition, we performed traditional morphometric analyses, meristic, hemipenis morphology and coloration pattern of H. gigas and H. melanogigas. Results. According to molecular and morphological characters, H. gigas is widely Submitted 19 May 2020 distributed throughout South America. We found no evidence to support that H. Accepted 9 September 2020 gigas and H. melanogigas species are distinct lineages, therefore, H. melanogigas is a Published 25 November 2020 junior synonym of H. -

Briefly Foreign Aid to Small Island Nations in Gests That Deserts May Be Among Those Return for Their Support Within the Ecosystems Most Affected
the buying of votes by promising to give Nations Environment Programme sug- Briefly foreign aid to small island nations in gests that deserts may be among those return for their support within the ecosystems most affected. Climatic commission. It seems, however, that pulses are more important than average these tactics are similar to those insti- conditions in desert ecosystems, and gated by Peter Scott, then head of WWF, because of this even moderate changes International to obtain the original moratorium on in precipitation and temperature can whaling. Evidence shows that many of have a severe impact. Contrary to the countries that voted in favour of the appearance, the 3.7 million km2 of the moratorium in 1982 had been offered world’s deserts provide a habitat for Hope for coral reefs help with providing suitable delegates many species, which will be adversely Many coral species are sensitive to rises and expenses. Scott’s biographer writes affected should the report’s projected in ocean temperature, and coral bleach- that China’s decision to join the IWC scenario of an increase in desert tem- ing events have increased in frequency and vote for the moratorium was influ- perature by 7˚C and a decrease in rain- in recent years. Now researchers have enced by a WWF promise to provide fall of 20% prove correct. However, shown that some corals may be able USD 1 million towards a panda reserve. deserts could also have a role to play to acclimatize to higher temperatures. Source: New Scientist (2006), 190(2556), in mitigating future environmental Experiments involving the hard coral 14. -

Checklist Reptile and Amphibian
To report sightings, contact: Natural Resources Coordinator 980-314-1119 www.parkandrec.com REPTILE AND AMPHIBIAN CHECKLIST Mecklenburg County, NC: 66 species Mole Salamanders ☐ Pickerel Frog ☐ Ground Skink (Scincella lateralis) ☐ Spotted Salamander (Rana (Lithobates) palustris) Whiptails (Ambystoma maculatum) ☐ Southern Leopard Frog ☐ Six-lined Racerunner ☐ Marbled Salamander (Rana (Lithobates) sphenocephala (Aspidoscelis sexlineata) (Ambystoma opacum) (sphenocephalus)) Nonvenomous Snakes Lungless Salamanders Snapping Turtles ☐ Eastern Worm Snake ☐ Dusky Salamander (Desmognathus fuscus) ☐ Common Snapping Turtle (Carphophis amoenus) ☐ Southern Two-lined Salamander (Chelydra serpentina) ☐ Scarlet Snake1 (Cemophora coccinea) (Eurycea cirrigera) Box and Water Turtles ☐ Black Racer (Coluber constrictor) ☐ Three-lined Salamander ☐ Northern Painted Turtle ☐ Ring-necked Snake (Eurycea guttolineata) (Chrysemys picta) (Diadophis punctatus) ☐ Spring Salamander ☐ Spotted Turtle2, 6 (Clemmys guttata) ☐ Corn Snake (Pantherophis guttatus) (Gyrinophilus porphyriticus) ☐ River Cooter (Pseudemys concinna) ☐ Rat Snake (Pantherophis alleghaniensis) ☐ Slimy Salamander (Plethodon glutinosus) ☐ Eastern Box Turtle (Terrapene carolina) ☐ Eastern Hognose Snake ☐ Mud Salamander (Pseudotriton montanus) ☐ Yellow-bellied Slider (Trachemys scripta) (Heterodon platirhinos) ☐ Red Salamander (Pseudotriton ruber) ☐ Red-eared Slider3 ☐ Mole Kingsnake Newts (Trachemys scripta elegans) (Lampropeltis calligaster) ☐ Red-spotted Newt Mud and Musk Turtles ☐ Eastern Kingsnake -

Snakes of South-East Asia Including Myanmar, Thailand, Malaysia, Singapore, Sumatra, Borneo, Java and Bali
A Naturalist’s Guide to the SNAKES OF SOUTH-EAST ASIA including Myanmar, Thailand, Malaysia, Singapore, Sumatra, Borneo, Java and Bali Indraneil Das First published in the United Kingdom in 2012 by Beaufoy Books n n 11 Blenheim Court, 316 Woodstock Road, Oxford OX2 7NS, England Contents www.johnbeaufoy.com 10 9 8 7 6 5 4 3 2 1 Introduction 4 Copyright © 2012 John Beaufoy Publishing Limited Copyright in text © Indraneil Das Snake Topography 4 Copyright in photographs © [to come] Dealing with Snake Bites 6 All rights reserved. No part of this publication may be reproduced, stored in a retrieval system or transmitted in any form or by any means, electronic, mechanical, photocopying, recording or otherwise, without the prior written permission of the publishers. About this Book 7 ISBN [to come] Glossary 8 Edited, designed and typeset by D & N Publishing, Baydon, Wiltshire, UK Printed and bound [to come] Species Accounts and Photographs 11 Checklist of South-East Asian Snakes 141 Dedication Nothing would have happened without the support of the folks at home: my wife, Genevieve V.A. Gee, and son, Rahul Das. To them, I dedicate this book. Further Reading 154 Acknowledgements 155 Index 157 Edited and designed by D & N Publishing, Baydon, Wiltshire, UK Printed and bound in Malaysia by Times Offset (M) Sdn. Bhd. n Introduction n n Snake Topography n INTRODUCTION Snakes form one of the major components of vertebrate fauna of South-East Asia. They feature prominently in folklore, mythology and other belief systems of the indigenous people of the region, and are of ecological and conservation value, some species supporting significant (albeit often illegal) economic activities (primarily, the snake-skin trade, but also sale of meat and other body parts that purportedly have medicinal properties). -

NHBSS 061 1G Hikida Fieldg
Book Review N$7+IST. BULL. S,$0 SOC. 61(1): 41–51, 2015 A Field Guide to the Reptiles of Thailand by Tanya Chan-ard, John W. K. Parr and Jarujin Nabhitabhata. Oxford University Press, New York, 2015. 344 pp. paper. ISBN: 9780199736492. 7KDLUHSWLOHVZHUHÀUVWH[WHQVLYHO\VWXGLHGE\WZRJUHDWKHUSHWRORJLVWV0DOFROP$UWKXU 6PLWKDQG(GZDUG+DUULVRQ7D\ORU7KHLUFRQWULEXWLRQVZHUHSXEOLVKHGDV6MITH (1931, 1935, 1943) and TAYLOR 5HFHQWO\RWKHUERRNVDERXWUHSWLOHVDQGDPSKLELDQV LQ7KDLODQGZHUHSXEOLVKHG HJ&HAN-ARD ET AL., 1999: COX ET AL DVZHOODVPDQ\ SDSHUV+RZHYHUWKHVHERRNVZHUHWD[RQRPLFVWXGLHVDQGQRWJXLGHVIRURUGLQDU\SHRSOH7ZR DGGLWLRQDOÀHOGJXLGHERRNVRQUHSWLOHVRUDPSKLELDQVDQGUHSWLOHVKDYHDOVREHHQSXEOLVKHG 0ANTHEY & GROSSMANN, 1997; DAS EXWWKHVHERRNVFRYHURQO\DSDUWRIWKHIDXQD The book under review is very well prepared and will help us know Thai reptiles better. 2QHRIWKHDXWKRUV-DUXMLQ1DEKLWDEKDWDZDVP\ROGIULHQGIRUPHUO\WKH'LUHFWRURI1DWXUDO +LVWRU\0XVHXPWKH1DWLRQDO6FLHQFH0XVHXP7KDLODQG+HZDVDQH[FHOOHQWQDWXUDOLVW DQGKDGH[WHQVLYHNQRZOHGJHDERXW7KDLDQLPDOVHVSHFLDOO\DPSKLELDQVDQGUHSWLOHV,Q ZHYLVLWHG.KDR6RL'DR:LOGOLIH6DQFWXDU\WRVXUYH\KHUSHWRIDXQD+HDGYLVHGXV WRGLJTXLFNO\DURXQGWKHUH:HFROOHFWHGIRXUVSHFLPHQVRIDibamusZKLFKZHGHVFULEHG DVDQHZVSHFLHVDibamus somsaki +ONDA ET AL 1RZ,DPYHU\JODGWRNQRZWKDW WKLVERRNZDVSXEOLVKHGE\KLPDQGKLVFROOHDJXHV8QIRUWXQDWHO\KHSDVVHGDZD\LQ +LVXQWLPHO\GHDWKPD\KDYHGHOD\HGWKHSXEOLFDWLRQRIWKLVERRN7KHERRNLQFOXGHVQHDUO\ DOOQDWLYHUHSWLOHV PRUHWKDQVSHFLHV LQ7KDLODQGDQGPRVWSLFWXUHVZHUHGUDZQZLWK H[FHOOHQWGHWDLO,WLVDYHU\JRRGÀHOGJXLGHIRULGHQWLÀFDWLRQRI7KDLUHSWLOHVIRUVWXGHQWV -

Borneo) in Two Different Ways
Contributions to Zoology, 78 (4) 141-147 (2009) Estimating the snake species richness of the Santubong Peninsula (Borneo) in two different ways Johan van Rooijen1, 2, 3 1 Zoological Museum Amsterdam, Mauritskade 61, 1092 AD Amsterdam, The Netherlands 2 Tulpentuin 313, 2272 EH Voorburg, The Netherlands 3 E-mail: [email protected] Key words: Chao I estimator, negative exponential function, rarefaction curve, Santubong Peninsula Borneo, snakes, species richness, Weibull function Abstract stantial investments in terms of search effort. This is particularly true for snakes which are hard to find (e.g. The distribution of Borneo’s species across the island is far Lloyd et al., 1968; Inger and Colwell, 1977; Hofer and from well-known. This is particularly true for snakes which are hard to find. Given the current rate of habitat destruction and Bersier, 2001; Orlov et al., 2003). As a consequence, consequent need for conservation strategies, more information estimation techniques are of interest when the intend- is required as to the species composition and richness of spe- ed objective is to assess species richness, an elemen- cific areas of potential conservation priority. An example is the tary criterion conservationists may use when identify- Santubong Peninsula, Sarawak, Malaysia, part of which has re- ing priority areas. One such estimation technique con- cently been gazetted as a National Park. In this paper, the snake species richness of the Santubong Peninsula is estimated on the sists of extrapolating the species accumulation curve. basis of data obtained during 450 survey-hours. Thirty-two spe- Species accumulation curves are regularly applied in cies were recorded. -

Snakes of the Everglades Agricultural Area1 Michelle L
CIR1462 Snakes of the Everglades Agricultural Area1 Michelle L. Casler, Elise V. Pearlstine, Frank J. Mazzotti, and Kenneth L. Krysko2 Background snakes are often escapees or are released deliberately and illegally by owners who can no longer care for them. Snakes are members of the vertebrate order Squamata However, there has been no documentation of these snakes (suborder Serpentes) and are most closely related to lizards breeding in the EAA (Tennant 1997). (suborder Sauria). All snakes are legless and have elongated trunks. They can be found in a variety of habitats and are able to climb trees; swim through streams, lakes, or oceans; Benefits of Snakes and move across sand or through leaf litter in a forest. Snakes are an important part of the environment and play Often secretive, they rely on scent rather than vision for a role in keeping the balance of nature. They aid in the social and predatory behaviors. A snake’s skull is highly control of rodents and invertebrates. Also, some snakes modified and has a great degree of flexibility, called cranial prey on other snakes. The Florida kingsnake (Lampropeltis kinesis, that allows it to swallow prey much larger than its getula floridana), for example, prefers snakes as prey and head. will even eat venomous species. Snakes also provide a food source for other animals such as birds and alligators. Of the 45 snake species (70 subspecies) that occur through- out Florida, 23 may be found in the Everglades Agricultural Snake Conservation Area (EAA). Of the 23, only four are venomous. The venomous species that may occur in the EAA are the coral Loss of habitat is the most significant problem facing many snake (Micrurus fulvius fulvius), Florida cottonmouth wildlife species in Florida, snakes included. -

Download Download
HAMADRYAD Vol. 27. No. 2. August, 2003 Date of issue: 31 August, 2003 ISSN 0972-205X CONTENTS T. -M. LEONG,L.L.GRISMER &MUMPUNI. Preliminary checklists of the herpetofauna of the Anambas and Natuna Islands (South China Sea) ..................................................165–174 T.-M. LEONG & C-F. LIM. The tadpole of Rana miopus Boulenger, 1918 from Peninsular Malaysia ...............175–178 N. D. RATHNAYAKE,N.D.HERATH,K.K.HEWAMATHES &S.JAYALATH. The thermal behaviour, diurnal activity pattern and body temperature of Varanus salvator in central Sri Lanka .........................179–184 B. TRIPATHY,B.PANDAV &R.C.PANIGRAHY. Hatching success and orientation in Lepidochelys olivacea (Eschscholtz, 1829) at Rushikulya Rookery, Orissa, India ......................................185–192 L. QUYET &T.ZIEGLER. First record of the Chinese crocodile lizard from outside of China: report on a population of Shinisaurus crocodilurus Ahl, 1930 from north-eastern Vietnam ..................193–199 O. S. G. PAUWELS,V.MAMONEKENE,P.DUMONT,W.R.BRANCH,M.BURGER &S.LAVOUÉ. Diet records for Crocodylus cataphractus (Reptilia: Crocodylidae) at Lake Divangui, Ogooué-Maritime Province, south-western Gabon......................................................200–204 A. M. BAUER. On the status of the name Oligodon taeniolatus (Jerdon, 1853) and its long-ignored senior synonym and secondary homonym, Oligodon taeniolatus (Daudin, 1803) ........................205–213 W. P. MCCORD,O.S.G.PAUWELS,R.BOUR,F.CHÉROT,J.IVERSON,P.C.H.PRITCHARD,K.THIRAKHUPT, W. KITIMASAK &T.BUNDHITWONGRUT. Chitra burmanica sensu Jaruthanin, 2002 (Testudines: Trionychidae): an unavailable name ............................................................214–216 V. GIRI,A.M.BAUER &N.CHATURVEDI. Notes on the distribution, natural history and variation of Hemidactylus giganteus Stoliczka, 1871 ................................................217–221 V. WALLACH.